Redundant Functions for Nap1 and Chz1 in H2A.Z Deposition
- PMID: 28883625
- PMCID: PMC5589762
- DOI: 10.1038/s41598-017-11003-8
Redundant Functions for Nap1 and Chz1 in H2A.Z Deposition
Abstract
H2A.Z is a histone H2A variant that contributes to transcriptional regulation, DNA damage response and limits heterochromatin spreading. In Saccharomyces cerevisiae, H2A.Z is deposited by the SWR-C complex, which relies on several histone chaperones including Nap1 and Chz1 to deliver H2A.Z-H2B dimers to SWR-C. However, the mechanisms by which Nap1 and Chz1 cooperate to bind H2A.Z and their contribution to H2A.Z deposition in chromatin is not well understood. Using structural modeling and molecular dynamics simulations, we identify a series of H2A.Z residues that form a chaperone-specific binding surface. Mutation of these residues revealed different surface requirements for Nap1 and Chz1 interaction with H2A.Z. Consistent with this result, we found that loss of Nap1 or Chz1 individually resulted in mild defects in H2A.Z deposition, but that deletion of both Nap1 and Chz1 resulted in a significant reduction of H2A.Z deposition at promoters and led to heterochromatin spreading. Together, our findings reveal unique H2A.Z surface dependences for Nap1 and Chz1 and a redundant role for these chaperones in H2A.Z deposition.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

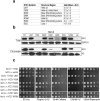


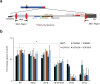
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

