Drug resistance in bacteria isolated from patients presenting with wounds at a non-profit Surgical Center in Phnom Penh, Cambodia from 2011-2013
- PMID: 28883936
- PMCID: PMC5526368
- DOI: 10.1186/s40794-015-0006-5
Drug resistance in bacteria isolated from patients presenting with wounds at a non-profit Surgical Center in Phnom Penh, Cambodia from 2011-2013
Abstract
Background: Emerging antibiotic resistance amongst clinically significant bacteria is a public health issue of increasing significance worldwide, but it is relatively uncharacterized in Cambodia. In this study we performed standard bacterial cultures on samples from wounds at a Non-Governmental-Organization (NGO) Hospital in Phnom Penh, Cambodia. Testing was performed to elucidate pathogenic bacteria causing wound infections and the antibiotic resistance profiles of bacterial isolates. All testing was performed at the Naval Medical Research Unit, No.2 (NAMRU-2) main laboratory in Phnom Penh, Cambodia.
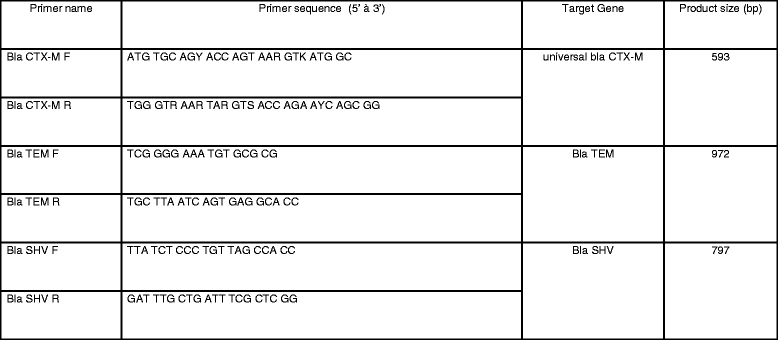
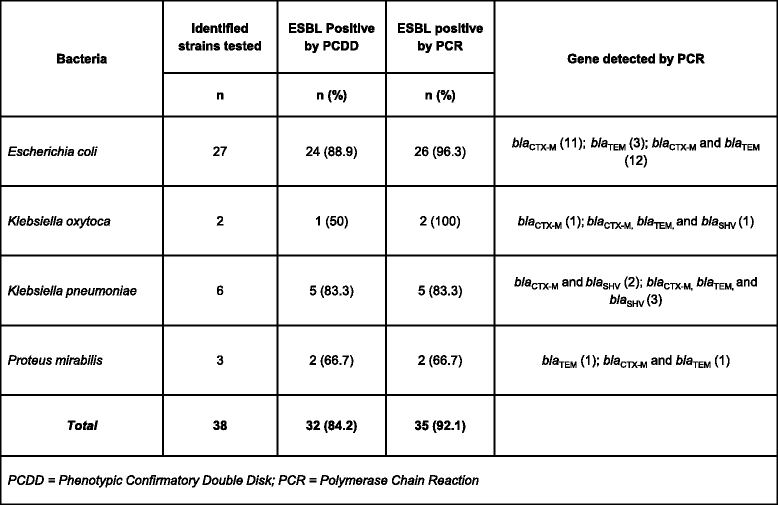
Methods: Between 2011-2013, a total of 251 specimens were collected from patients at the NGO hospital and analyzed for bacterial infection by standard bacterial cultures techniques. Specimens were all from wounds and anonymous. No specific clinical information accompanied the submitted specimens. Antibiotic susceptibility testing, and phenotypic testing for extended-spectrum beta-lactamase (ESBL) were performed and reported based on CLSI guidelines. Further genetic testing for CTX-M, TEM and SHV ESBLs was accomplished using PCR.
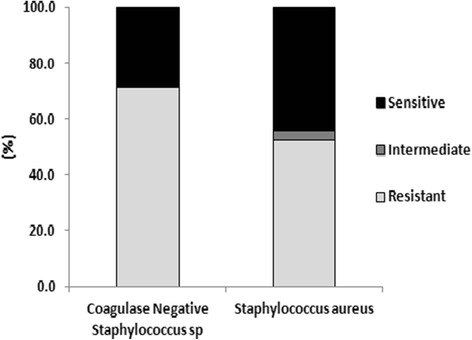
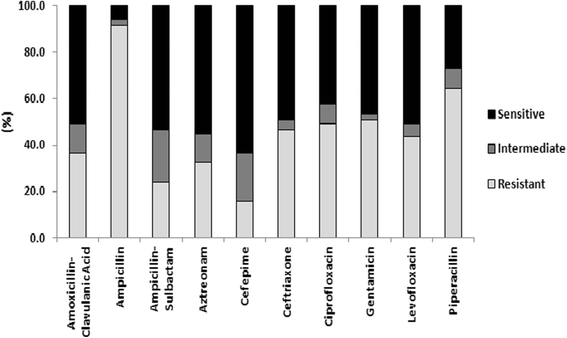
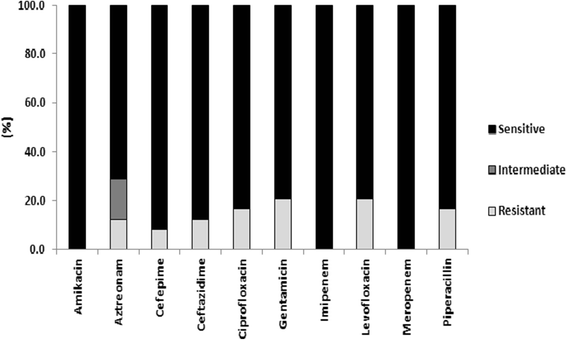
Results: One-hundred and seventy-six specimens were positive following bacterial culture (70 %). Staphlycoccus aureus was the most frequently isolated bacteria. Antibiotic drug resistance testing revealed that 52.5 % of Staphlycoccus aureus isolates were oxacillin resistant. For Escherichia coli isolates, 63.9 % were ciprofloxacin and levofloxacin resistant and 96 % were ESBL producers. Resistance to meropenem and imipenem was observed in one of three Acinetobacter spp isolates.
Conclusions: This study is the first of its kind detailing the antibiotic resistance profiles of pathogenic bacteria causing wound infections at a single surgical hospital in Cambodia. The reported findings of this study demonstrate significant antibiotic resistance in bacteria from injured patients and should serve to guide treatment modalities in Cambodia.
Keywords: Bacteria; Cambodia; Microbial drug resistance; Wound infections.
Figures
Similar articles
-
Prevalence and distribution of beta-lactamase coding genes in third-generation cephalosporin-resistant Enterobacteriaceae from bloodstream infections in Cambodia.Eur J Clin Microbiol Infect Dis. 2015 Jun;34(6):1223-9. doi: 10.1007/s10096-015-2350-9. Epub 2015 Feb 26. Eur J Clin Microbiol Infect Dis. 2015. PMID: 25717021 Free PMC article.
-
Bloodstream infection among adults in Phnom Penh, Cambodia: key pathogens and resistance patterns.PLoS One. 2013;8(3):e59775. doi: 10.1371/journal.pone.0059775. Epub 2013 Mar 29. PLoS One. 2013. PMID: 23555777 Free PMC article.
-
Molecular characteristics and antibiotic resistance profiles of Escherichia coli strains isolated from urinary tract infections in children admitted to children's referral hospital of Qom, Iran.Ann Ig. 2019 May-Jun;31(3):252-262. doi: 10.7416/ai.2019.2288. Ann Ig. 2019. PMID: 31069370
-
[Investigation of beta-lactamase genes and clonal relationship among the extended-spectrum beta-lactamase producing nosocomial Escherichia coli isolates].Mikrobiyol Bul. 2015 Jan;49(1):15-25. doi: 10.5578/mb.8437. Mikrobiyol Bul. 2015. PMID: 25706727 Turkish.
-
Distribution of blaCTX - M , blaTEM , blaSHV and blaOXA genes in Extended-spectrum-β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan.Antimicrob Resist Infect Control. 2019 May 22;8:80. doi: 10.1186/s13756-019-0536-0. eCollection 2019. Antimicrob Resist Infect Control. 2019. PMID: 31139363 Free PMC article.
Cited by
-
Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections.PLoS One. 2017 Jul 18;12(7):e0179245. doi: 10.1371/journal.pone.0179245. eCollection 2017. PLoS One. 2017. PMID: 28719657 Free PMC article.
-
Public knowledge, attitudes and practices surrounding antibiotic use and resistance in Cambodia.JAC Antimicrob Resist. 2021 Feb 3;3(1):dlaa115. doi: 10.1093/jacamr/dlaa115. eCollection 2021 Mar. JAC Antimicrob Resist. 2021. PMID: 34223067 Free PMC article.
-
An Observational Study of Sepsis in Takeo Province Cambodia: An in-depth examination of pathogens causing severe infections.PLoS Negl Trop Dis. 2020 Aug 17;14(8):e0008381. doi: 10.1371/journal.pntd.0008381. eCollection 2020 Aug. PLoS Negl Trop Dis. 2020. PMID: 32804954 Free PMC article.
-
Methicillin resistant Staphylococcus aureus isolated from surgical patients in Cambodia over a 10-year period.Trop Doct. 2023 Jul;53(3):347-351. doi: 10.1177/00494755231174261. Epub 2023 May 8. Trop Doct. 2023. PMID: 37157819 Free PMC article.
-
Epidemiology of Extended-Spectrum Beta-Lactamase and Carbapenemase-Producing Enterobacterales in the Greater Mekong Subregion: A Systematic-Review and Meta-Analysis of Risk Factors Associated With Extended-Spectrum Beta-Lactamase and Carbapenemase Isolation.Front Microbiol. 2021 Nov 26;12:695027. doi: 10.3389/fmicb.2021.695027. eCollection 2021. Front Microbiol. 2021. PMID: 34899618 Free PMC article. Review.
References
-
- van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother. 2012;67:2090–100. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

