Effects of storage temperature on the quantity and integrity of genomic DNA extracted from mice tissues: A comparison of recovery methods
- PMID: 28884076
- PMCID: PMC5579564
- DOI: 10.4314/ovj.v7i3.7
Effects of storage temperature on the quantity and integrity of genomic DNA extracted from mice tissues: A comparison of recovery methods
Abstract
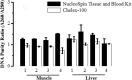
Efficient extraction of genomic DNA (gDNA) from biological materials found in harsh environments is the first step for successful forensic DNA profiling. This study aimed to evaluate two methods for DNA recovery from animal tissues (livers, muscles), focusing on the best storage temperature for DNA yield in term of quality, quantity, and integrity for use in several downstream molecular techniques. Six male Swiss albino mice were sacrificed, liver and muscle tissues (n=32) were then harvested and stored for one week in different temperatures, -20°C, 4°C, 25°C and 40°C. The conditioned animal tissues were used for DNA extraction by Chelex-100 method or NucleoSpinC Blood and Tissue kit. The extracted gDNA was visualized on 1.5% agarose gel electrophoresis to determine the quality of gDNA and analysed spectrophotometrically to determine the DNA concentration and the purity. Both methods, Chelex-100 and NucleoSpin Blood and Tissue kit found to be appropriate for yielding high quantity of gDNA, with the Chelex 100 method yielding a greater quantity (P < 0.045) than the kit. At -20°C, 4°C, and 25°C temperatures, the concentration of DNA yield was numerically lower than at 40°C. The NucleoSpinC Blood and Tissue kit produced a higher (P=0.031) purity product than the Chelex-100 method, particularly for muscle tissues. The Chelex-100 method is cheap, fast, effective, and is a crucial tool for yielding DNA from animal tissues (livers, muscles) exposed to harsh environment with little limitations.
Keywords: DNA degradation; DNA extraction; DNA profiling; Purity; Temperature.
Figures


References
-
- Bogas V, Balsa F, Carvalho M, Anjos M. J, Pinheiro M. F, Corte-Real F. Comparison of four DNA extraction methods for forensic application. Forensic Science International: Genetics Supplement Series. 2011;3(1):e194–e195.
-
- Dhaliwal A. DNA Extraction and Purification. MATER METHODS [Online] 2013. Available at: https://www.labome.com/method/DNA-Extraction-and-Purification.html .
-
- Ebuehi O.A.T, Amode M, Balogun A, Fowora A. Postmortem Time Affects Brain, Liver, Kidney and Heart DNA in Male Rat. Am. J. Biochem. 2015;5(1):1–5.
-
- El-Harouny M, El-Dakroory S, Attalla S, Hasan N, Hassab El-Nabi S.E. The relationship between postmortem interval and DNA degradation in different tissues of drowned rats. Mansoura J. Forensic Med. Clin. Toxicol. 2008;16(2):45–61.
LinkOut - more resources
Full Text Sources
Other Literature Sources
