Inhibition of DYRK1A disrupts neural lineage specificationin human pluripotent stem cells
- PMID: 28884684
- PMCID: PMC5656431
- DOI: 10.7554/eLife.24502
Inhibition of DYRK1A disrupts neural lineage specificationin human pluripotent stem cells
Abstract
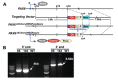
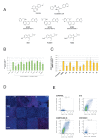
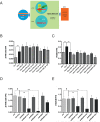
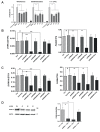
Genetic analysis has revealed that the dual specificity protein kinase DYRK1A has multiple roles in the development of the central nervous system. Increased DYRK1A gene dosage, such as occurs in Down syndrome, is known to affect neural progenitor cell differentiation, while haploinsufficiency of DYRK1A is associated with severe microcephaly. Using a set of known and newly synthesized DYRK1A inhibitors, along with CRISPR-mediated gene activation and shRNA knockdown of DYRK1A, we show here that chemical inhibition or genetic knockdown of DYRK1A interferes with neural specification of human pluripotent stem cells, a process equating to the earliest stage of human brain development. Specifically, DYRK1A inhibition insulates the self-renewing subpopulation of human pluripotent stem cells from powerful signals that drive neural induction. Our results suggest a novel mechanism for the disruptive effects of the absence or haploinsufficiency of DYRK1A on early mammalian development, and reveal a requirement for DYRK1A in the acquisition of competence for differentiation in human pluripotent stem cells.
Keywords: Homo sapiens; developmental biology; none; pluripotent; stem cell; stem cells.
Conflict of interest statement
Reviewing editor,
No competing interests declared.
Figures





References
-
- Briggs JA, Sun J, Shepherd J, Ovchinnikov DA, Chung TL, Nayler SP, Kao LP, Morrow CA, Thakar NY, Soo SY, Peura T, Grimmond S, Wolvetang EJ. Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells. 2013;31:467–478. doi: 10.1002/stem.1297. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

