Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease
- PMID: 28885228
- PMCID: PMC6461205
- DOI: 10.1097/MIB.0000000000001246
Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease
Abstract
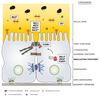
Background: Intestinal barrier defects are common in patients with inflammatory bowel disease (IBD). To identify which components could underlie these changes, we performed an in-depth analysis of epithelial barrier genes in IBD.
Methods: A set of 128 intestinal barrier genes was selected. Polygenic risk scores were generated based on selected barrier gene variants that were associated with Crohn's disease (CD) or ulcerative colitis (UC) in our study. Gene expression was analyzed using microarray and quantitative reverse transcription polymerase chain reaction. Influence of barrier gene variants on expression was studied by cis-expression quantitative trait loci mapping and comparing patients with low- and high-risk scores.
Results: Barrier risk scores were significantly higher in patients with IBD than controls. At single-gene level, the associated barrier single-nucleotide polymorphisms were most significantly enriched in PTGER4 for CD and HNF4A for UC. As a group, the regulating proteins were most enriched for CD and UC. Expression analysis showed that many epithelial barrier genes were significantly dysregulated in active CD and UC, with overrepresentation of mucus layer genes. In uninflamed CD ileum and IBD colon, most barrier gene levels restored to normal, except for MUC1 and MUC4 that remained persistently increased compared with controls. Expression levels did not depend on cis-regulatory variants nor combined genetic risk.
Conclusions: We found genetic and transcriptomic dysregulations of key epithelial barrier genes and components in IBD. Of these, we believe that mucus genes, in particular MUC1 and MUC4, play an essential role in the pathogenesis of IBD and could represent interesting targets for treatment.
Figures
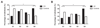


References
-
- Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54 e42. quiz e30. - PubMed
-
- Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. - PubMed
-
- Rampton D, Shanahan F. Fast facts inflammatory bowel disease. 3. ed. Abingdon, Oxford: Health Press Ltd; 2008.
-
- McGuckin MA, Eri R, Simms LA, et al. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

