A study of the structural properties of sites modified by the O-linked 6-N-acetylglucosamine transferase
- PMID: 28886091
- PMCID: PMC5590929
- DOI: 10.1371/journal.pone.0184405
A study of the structural properties of sites modified by the O-linked 6-N-acetylglucosamine transferase
Erratum in
-
Correction: A study of the structural properties of sites modified by the O-linked 6-N-acetylglucosamine transferase.PLoS One. 2017 Dec 27;12(12):e0190461. doi: 10.1371/journal.pone.0190461. eCollection 2017. PLoS One. 2017. PMID: 29281737 Free PMC article.
Abstract
Protein O-GlcNAcylation (O-GlcNAc) is an essential post-translational modification (PTM) in higher eukaryotes. The O-linked β-N-acetylglucosamine transferase (OGT), targets specific Serines and Threonines (S/T) in intracellular proteins. However, unlike phosphorylation, fewer than 25% of known O-GlcNAc sites match a clear sequence pattern. Accordingly, the three-dimensional structures of O-GlcNAc sites were characterised to investigate the role of structure in molecular recognition. From 1,584 O-GlcNAc sites in 620 proteins, 143 were mapped to protein structures determined by X-ray crystallography. The modified S/T were 1.7 times more likely to be annotated in the REM465 field which defines missing residues in a protein structure, while 7 O-GlcNAc sites were solvent inaccessible and unlikely to be targeted by OGT. 132 sites with complete backbone atoms clustered into 10 groups, but these were indistinguishable from clusters from unmodified S/T. This suggests there is no prevalent three-dimensional motif for OGT recognition. Predicted features from the 620 proteins were compared to unmodified S/T in O-GlcNAcylated proteins and globular proteins. The Jpred4 predicted secondary structure shows that modified S/T were more likely to be coils. 5/6 methods to predict intrinsic disorder indicated O-GlcNAcylated S/T to be significantly more disordered than unmodified S/T. Although the analysis did not find a pattern in the site three-dimensional structure, it revealed the residues around the modification site are likely to be disordered and suggests a potential role of secondary structure elements in OGT site recognition.
Conflict of interest statement
Figures

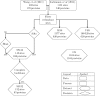

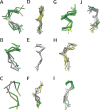

References
-
- Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97: 5735–9. doi: 10.1073/pnas.100471497 - DOI - PMC - PubMed
-
- O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-Dependent X-Chromosome-Linked Protein Glycosylation Is a Requisite Modification in Somatic Cell Function and Embryo Viability. Mol Cell Biol. 2004;24: 1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004 - DOI - PMC - PubMed
-
- Buse MG. Hexosamines, insulin resistance, and the complications of diabetes: current status. Am J Physiol Endocrinol Metab. 2006;290: E1–E8. doi: 10.1152/ajpendo.00329.2005 - DOI - PMC - PubMed
-
- Abdel Rahman AM, Ryczko M, Pawling J, Dennis JW. Probing the hexosamine biosynthetic pathway in human tumor cells by multitargeted tandem mass spectrometry. ACS Chem Biol. 2013;8: 2053–62. doi: 10.1021/cb4004173 - DOI - PubMed
-
- Liu J, Marchase RB, Chatham JC. Increased O-GlcNAc levels during reperfusion lead to improved functional recovery and reduced calpain proteolysis. Am J Physiol Heart Circ Physiol. 2007;293: H1391–9. doi: 10.1152/ajpheart.00285.2007 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

