The plant RNA polymerase II elongation complex: A hub coordinating transcript elongation and mRNA processing
- PMID: 28886274
- PMCID: PMC5834219
- DOI: 10.1080/21541264.2017.1356902
The plant RNA polymerase II elongation complex: A hub coordinating transcript elongation and mRNA processing
Abstract
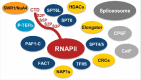
Characterisation of the Arabidopsis RNA polymerase II (RNAPII) elongation complex revealed an assembly of a conserved set of transcript elongation factors associated with chromatin remodellers, histone modifiers as well as with various pre-mRNA splicing and polyadenylation factors. Therefore, transcribing RNAPII streamlines the processes of mRNA synthesis and processing in plants.
Keywords: RNA polymerase II; chromatin; polyadenylation; splicing; transcript elongation.
Figures

Similar articles
-
The Composition of the Arabidopsis RNA Polymerase II Transcript Elongation Complex Reveals the Interplay between Elongation and mRNA Processing Factors.Plant Cell. 2017 Apr;29(4):854-870. doi: 10.1105/tpc.16.00735. Epub 2017 Mar 28. Plant Cell. 2017. PMID: 28351991 Free PMC article.
-
Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis.J Exp Bot. 2014 May;65(8):1973-80. doi: 10.1093/jxb/eru085. Epub 2014 Mar 6. J Exp Bot. 2014. PMID: 24604736 Review.
-
Elongator and SPT4/SPT5 complexes as proxy to study RNA polymerase II transcript elongation control of plant development.Proteomics. 2014 Oct;14(19):2109-14. doi: 10.1002/pmic.201400024. Epub 2014 May 22. Proteomics. 2014. PMID: 24733746
-
Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing.Proc Natl Acad Sci U S A. 2015 Dec 1;112(48):14840-5. doi: 10.1073/pnas.1506760112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578803 Free PMC article.
-
Transcript elongation factors: shaping transcriptomes after transcript initiation.Trends Plant Sci. 2014 Nov;19(11):717-26. doi: 10.1016/j.tplants.2014.07.002. Epub 2014 Aug 11. Trends Plant Sci. 2014. PMID: 25131948 Review.
Cited by
-
TFIIS is required for reproductive development and thermal adaptation in barley.Plant Cell Rep. 2024 Oct 10;43(11):260. doi: 10.1007/s00299-024-03345-1. Plant Cell Rep. 2024. PMID: 39390135 Free PMC article.
-
The FACT Histone Chaperone: Tuning Gene Transcription in the Chromatin Context to Modulate Plant Growth and Development.Front Plant Sci. 2020 Feb 19;11:85. doi: 10.3389/fpls.2020.00085. eCollection 2020. Front Plant Sci. 2020. PMID: 32140163 Free PMC article. Review.
-
The chromatin remodeler ZmCHB101 impacts expression of osmotic stress-responsive genes in maize.Plant Mol Biol. 2018 Jul;97(4-5):451-465. doi: 10.1007/s11103-018-0751-8. Epub 2018 Jun 28. Plant Mol Biol. 2018. PMID: 29956114
-
The Physcomitrella patens chromatin adaptor PpMRG1 interacts with H3K36me3 and regulates light-responsive alternative splicing.Plant Physiol. 2021 Apr 2;185(3):1229-1241. doi: 10.1093/plphys/kiaa103. Plant Physiol. 2021. PMID: 33793927 Free PMC article.
-
Alternative Splicing and Protein Diversity: Plants Versus Animals.Front Plant Sci. 2019 Jun 12;10:708. doi: 10.3389/fpls.2019.00708. eCollection 2019. Front Plant Sci. 2019. PMID: 31244866 Free PMC article. Review.
References
-
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167-77. http://doi.org/10.1038/nrm3953. PMID:25693130 - DOI - PMC - PubMed
-
- Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and the long of it. Genes Dev. 2004;18:2437-68. http://doi.org/10.1101/gad.1235904. PMID:15489290 - DOI - PubMed
-
- Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 2013;27:1079-88. http://doi.org/10.1101/gad.215137.113. PMID:23699407 - DOI - PMC - PubMed
-
- Van Lijsebettens M, Grasser KD. Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 2014;19:717-26. http://doi.org/10.1016/j.tplants.2014.07.002. PMID:25131948 - DOI - PubMed
-
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979-92. http://doi.org/10.1128/MCB.22.20.6979-6992.2002. PMID:12242279 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
