A New Diagnostic Resource for Ceratitis capitata Strain Identification Based on QTL Mapping
- PMID: 28889103
- PMCID: PMC5677166
- DOI: 10.1534/g3.117.300169
A New Diagnostic Resource for Ceratitis capitata Strain Identification Based on QTL Mapping
Abstract
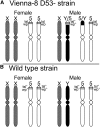
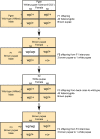
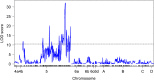
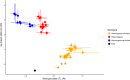
The Mediterranean fruit fly Ceratitis capitata (Wiedemann) is a destructive agricultural pest and the subject of exclusion efforts in many countries. Suppression and eradication of invasive populations to prevent its establishment is facilitated by the release of sterile males using the sterile insect technique (SIT). In SIT release areas, it is critical to accurately discriminate between released sterile males and wild individuals to detect extremely rare invasive individuals in areas inundated with millions of sterile male flies. Current methods for discrimination exist but are not always definitive, and a more reliable method is necessary. To address this, we developed a genotyping assay that can be used to discriminate between sterile males from the SIT strain and wild individuals. This was achieved by identifying single nucleotide polymorphisms (SNPs) linked to the maintained traits that facilitate male-only releases, white pupae (wp) and temperature-sensitive lethal (tsl), via QTL mapping. This resulted in the identification of one SNP that was in near-perfect linkage disequilibrium between genotype at this locus and the pupal color phenotype. Medfly from many SIT colonies and wild individuals from across its geographic range were genotyped for this locus, and results show its consistency in identifying SIT flies. In addition, linkage and QTL mapping of wp and tsl have larger impacts as they can serve as foundational tools to identify the genetic basis of traits that facilitate the separation of males from female flies, which can be used to develop SIT programs in related species.
Keywords: QTL; diagnostics; genome-wide genotyping; linkage mapping; medfly; sterile insect technique.
Copyright © 2017 Sim et al.
Figures


References
-
- Andrewartha H. G., Birch L. C., 1960. Some recent contributions to the study of the distribution and abundance of insects. Annu. Rev. Entomol. 5: 219–242.
-
- Barr N. B., 2009. Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J. Econ. Entomol. 102: 401–411. - PubMed
-
- Bassett A., Liu J. L., 2014. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69: 128–136. - PubMed
-
- Broman K. W., Wu H., Sen S., Churchill G., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
