An NF-κB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function
- PMID: 28889947
- PMCID: PMC5679261
- DOI: 10.1016/j.immuni.2017.08.010
An NF-κB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function
Abstract
Both conventional T (Tconv) cells and regulatory T (Treg) cells are activated through ligation of the T cell receptor (TCR) complex, leading to the induction of the transcription factor NF-κB. In Tconv cells, NF-κB regulates expression of genes essential for T cell activation, proliferation, and function. However the role of NF-κB in Treg function remains unclear. We conditionally deleted canonical NF-κB members p65 and c-Rel in developing and mature Treg cells and found they have unique but partially redundant roles. c-Rel was critical for thymic Treg development while p65 was essential for mature Treg identity and maintenance of immune tolerance. Transcriptome and NF-κB p65 binding analyses demonstrated a lineage specific, NF-κB-dependent transcriptional program, enabled by enhanced chromatin accessibility. These dual roles of canonical NF-κB in Tconv and Treg cells highlight the functional plasticity of the NF-κB signaling pathway and underscores the need for more selective strategies to therapeutically target NF-κB.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

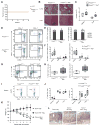



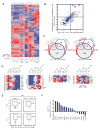

Comment in
-
Driving Rel-iant Tregs toward an Identity Crisis.Immunity. 2017 Sep 19;47(3):391-393. doi: 10.1016/j.immuni.2017.08.014. Immunity. 2017. PMID: 28930651
References
-
- Deenick EK, Elford AR, Pellegrini M, Hall H, Mak TW, Ohashi PS. c-Rel but not NF-kappaB1 is important for T regulatory cell development. Eur J Immunol. 2010;40:677–681. - PubMed
-
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

