Ionotropic Receptors Mediate Drosophila Oviposition Preference through Sour Gustatory Receptor Neurons
- PMID: 28889974
- PMCID: PMC5680077
- DOI: 10.1016/j.cub.2017.08.003
Ionotropic Receptors Mediate Drosophila Oviposition Preference through Sour Gustatory Receptor Neurons
Abstract
Carboxylic acids are present in many foods, being especially abundant in fruits. Yet, relatively little is known about how acids are detected by gustatory systems and whether they have a potential role in nutrition or provide other health benefits. Here we identify sour gustatory receptor neurons (GRNs) in tarsal taste sensilla of Drosophila melanogaster. We find that most tarsal sensilla harbor a sour GRN that is specifically activated by carboxylic and mineral acids but does not respond to sweet- and bitter-tasting chemicals or salt. One pair of taste sensilla features two GRNs that respond only to a subset of carboxylic acids and high concentrations of salt. All sour GRNs prominently express two Ionotropic Receptor (IR) genes, IR76b and IR25a, and we show that both these genes are necessary for the detection of acids. Furthermore, we establish that IR25a and IR76b are essential in sour GRNs of females for oviposition preference on acid-containing food. Our investigations reveal that acids activate a unique set of taste cells largely dedicated to sour taste, and they indicate that both pH/proton concentration and the structure of carboxylic acids contribute to sour GRN activation. Together, our studies provide new insights into the cellular and molecular basis of sour taste.
Keywords: Drosophila melanogaster; IR; IR25a; IR76b; carboxylic acid; gustatory receptor; gustatory receptor neuron; ionotropic receptor; oviposition; sour taste receptor.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
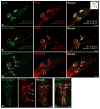
Immunostaining of fifth tarsal segment of a w1118/w1118; IR25a-GAL4/ IR76b-QF UAS-mCD8GFP; QUAS-mtdTomato-3XHA/+ fly. IR25a and IR76b are broadly co-expressed in a large number of tarsal GRNs. Inset shows schematic drawing of chemosensory sensilla in the fifth tarsal segment.
Immunostaining of the fifth tarsal segment of a w1118/w1118; IR76b-QF UAS-mCD8GFP/ Gr33a-GAL4, Gr64f-GAL4; QUAS-mtdTomato-3XHA/+ fly. Bitter and sweet GRNs express IR76b, but each sensillum harbors one (or two) additional GRN(s) that is not labeled by bitter and sweet Gr-GAL4 drivers. Arrow indicates the non-sweet and non-bitter neurons. Numbers in (A) and (B) represent the average number of labeled GRNs of the three taste sensilla: 5b and 5s+5v (Mean ± SEM, 6≦n≦8). Neurons of the 5a sensillum (dotted oval) respond to pheromones and were excluded in the count. Scale bar represents 10 μM.
Endogenous IR25a, visualized by anti IR25a antibody, and IR76b-GAL4 are co-expressed in IR76bonly GRNs of the foreleg: Genotype: w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-GCaMP6m; Gr64f-LexA/LexAop-GAL80. Note that the bitter GRN of the 5b sensillum (*) does not express Gr66-LexA.
Immunostaining of the ventral nerve cord in the thorax of a w1118/w1118; IR76b-QF UAS-mCD8GFP/ Gr33a-GAL4 Gr64f-GAL4; QUAS-mtdTomato-3XHA/+ fly. Tarsal projections of GRNs expressing IR76b (red) terminate predominantly in the thoracic ganglion, while most tarsal projections of GRNs expressing bitter Gr genes and a subset of GRNs expressing sweet Gr genes (green) bypass the ganglion to terminate in the SEZ. f=foreleg, m=midleg, h=hindleg, w=wing
Projections of tarsal and wing IR76bonly GRNs predominantly terminate in the thoracic ganglion. Genotype: w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-GCaMP6m; Gr64f-LexA/LexAop-GAL80.
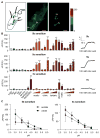
Schematic image of chemosensory sensilla in the fifth segment of tarsi (left), and live image of IR76bonly neuron after stimulation with 100mM acetic acid (middle). Relative fluorescence change (ΔF) is visualized in the right image. The scale bar represents 10μm. * indicates bitter GRN in 5b sensilla, which does not expressed Gr66a-LexA.
Ca2+ response (ΔF/F(%)) of IR76bonly GRNs after stimulation with indicated ligands. Response profile are similar for the 5b (top) and 5s (middle) associated sour GRNs (i.e. IR76bonly). 5v sensilla (bottom) harbor two similarly tuned IR76bonly GRNs that respond only to malic and glycolic acid (at high concentration), as well as to 500 mM NaCl. Representative traces to 100 mM citric acid are shown on the right of each graph. Amino acid (aa) mix of 40 mM contains all 20 amino acids (each at 2mM). Acetic acid concentrations: 10 mM, 100 mM, 500 mM; Citric and tartaric acid concentrations: 1 mM, 10 mM, 100 mM; Malic and glycolic acid concentration: 100mM; HCl concentrations: 1 mM, 10 mM, 25 mM, 50 mM. Data shown as mean and error bar indicate SEM, Student’s t test, *p<0.05, **p<0.01, ***p<0.001, 6≤n≤11.
Ca2+ response of IR76bonly GRNs in 5b and 5s sensilla stimulated with 100 mM acetate and citrate buffered solutions. 6≤n≤10.
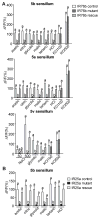
Absence of IR76b leads to a complete loss of sour GRN responses, which is rescued in the presence of an IR76b transgene, expressed under the control of IR76b-GAL4. Genotypes: w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-GCaMP6m; Gr64f-LexA/ LexAop-Gal80 (IR76b control), w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-GCaMP6m; Gr64f-LexA IR76b2/IR76b1 LexAop-GAL80 (IR76b mutant), w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-IR76b UAS-GCaMP6m; Gr64f-LexA IR76b2/ IR76b1 LexAop-GAL80 (IR76b rescue).
Absence of IR25a leads to a complete loss of sour GRN responses, which is rescued in the presence of an IR25a transgene, expressed under the control of IR25a-GAL4. Note that because the IR25a-Gal4 and Gr66a-LexA transgenes and IR25a2 are all located on the second chromosome, rescue experiments could not be carried out with the GAL80 suppressor transgenes. Consequently, only the 5b sensillum was analyzed, because the neurons in the 5v and the 5s sensilla are in too close proximity for unequivocal identification. Genotypes: w1118/w1118; IR25a-GAL4/+; UAS-GcaMP6m/+ (IR25a control), w1118/w1118; IR25a-GAL4 IR25a2/IR25a2;UAS-GCaMP6m/+ (IR25a mutant), w1118/w1118; IR25a-Gal4 IR25a2/IR25a2 UAS-IR25a; UAS-GCaMP6m /+ (IR25a rescue).
Concentrations used: 100 mM for acetic acid, citric acid and tartaric acid; 10 mM for glycolic acid and malic acid; 50 mM for HCl and NaCl; and 100 and 500 mM for KCl. All data are shown as mean and error bar indicate SEM, one-way ANOVA with post hoc Bonferroni correction, different letters indicate significant difference with p<0.05, 6≤n≤12.

Representative example of female oviposition preference on acid containing sugar-agar: the majority of eggs are laid on the acid containing half of the plate (left). w1118 females have a strong, dose dependent preference for oviposition on acid containing sugar-agar (right; see also Figure S4).
Silencing of all IR76b-GAL4 GRNs or sour (i.e. IR76bonly) GRNs abolishes acid preference, while silencing only the sweet and bitter GRNs has no effect. Genotypes: w1118/w1118 (WT), w1118/w1118; UAS-Kir2.1/+ (UAS control), IR76b-GAL4/UAS-Kir2.1 (Silencing IR76b GRNs), w1118/w1118; IR76b-GAL4 Gr66a-LexA/UAS-Kir2.1; Gr64f-LexA/LexAop-GAL8o (Silencing sour GRNs), w1118/w1118; Gr33a-GAL4 Gr64f-GAL4/UAS-Kir2.1 (Silencing bitter/sweet GRNs).
All major taste organs contribute to ovipositon preference for acetic acid (left), but removal of the forelegs has a more severe effect than removal of any other taste organ. The loss of oviposition preference for citric acid after removal of any specific taste organ is less pronounced, and significant reduction is only observed when the forelegs were removed. Genotype: w1118 (see also Figure S5)
Data shown as mean and error bar indicate SEM, Student t test for A), ***p<0.001, 20≤n≤30. One-way ANOVA with post hoc Bonferroni correction for B) and C), different letters indicate significant difference with p<0.05, 23≤n≤39
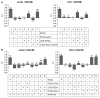
Females lacking IR76b lost preference for oviposition on acid containing sugar-agar, a phenotype that can be rescued by expressing IR76b under the control of IR76b-GAL4. Note that oviposition preference is also restored when IR76b is only expressed in sour GRNs (lane 7). Genotypes: IR76b+ control (w1118/w1118 lane1); w1118/w1118; IR76b1/ IR76b1 (2), w1118/w1118; IR76b-GAL4/+; IR76b1/IR76b1 (3), w1118/w1118; UAS-IR76b/+; IR76b1/IR76b1 (4), w1118/w1118; IR76b-GAL4/UAS-IR76b; IR76b1/IR76b1 (5), w1118/w1118; IR76b-GAL4 Gr66a- LexA/+; Gr64f-LexA IR76b2/ R76b1 LexAop-GAL8o (6) w1118/w1118; IR76b-GAL4 Gr66a- LexA/UAS-IR76b; Gr64f-LexA IR76b2/ R76b1 LexAop-GAL8o (7)
Females lacking IR25a lost preference for oviposition on acid containing sugar-agar. This phenotype was not rescued by expression of a UAS-IR25a gene under the control of the IR25a-GAL4 driver (lane 5), but it was rescued when expressed under the control of IR76b-GAL4 (Lanes 8 and 9; see text). Genotypes: IR25a+ control (w1118/w1118 lane 1), w1118/w1118;IR25a2/IR25a2 (2), w1118/w1118; IR25a-GAL4/+; IR25a2/IR25a2 (3), w1118/w1118; UAS-IR25a/+; IR25a2/IR25a2 (4), w1118/w1118; IR25a-GAL4 IR25a2/IR25a2 UAS-IR25a (5), w1118/w1118; IR76b-GAL4 IR25a2/IR25a2 (6), w1118/w1118; IR76b-GAL4 IR25a2/IR25a2 antenna removed (7), w1118/w1118; IR76b-GAL4 IR25a2/IR25a2 UAS-IR25a (8), w1118/w1118; IR76b-GAL4 IR25a2/IR25a2 UAS-IR25a antenna removed (9).
All data are shown as mean and error bar indicate SEM, one-way ANOVA with post hoc Bonferroni correction, different letters indicate significant difference with p<0.05, 20≤n≤39.
References
-
- Ramos Da Conceicao Neta ER, Johanningsmeier SD, McFeeters RF. The chemistry and physiology of sour taste--a review. J Food Sci. 2007;72:R33–38. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

