Rethinking HSF1 in Stress, Development, and Organismal Health
- PMID: 28890254
- PMCID: PMC5696061
- DOI: 10.1016/j.tcb.2017.08.002
Rethinking HSF1 in Stress, Development, and Organismal Health
Abstract
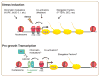
The heat shock response (HSR) was originally discovered as a transcriptional response to elevated temperature shock and led to the identification of heat shock proteins and heat shock factor 1 (HSF1). Since then HSF1 has been shown to be important for combating other forms of environmental perturbations as well as genetic variations that cause proteotoxic stress. The HSR has long been thought to be an absolute response to conditions of cell stress and the primary mechanism by which HSF1 promotes organismal health by preventing protein aggregation and subsequent proteome imbalance. Accumulating evidence now shows that HSF1, the central player in the HSR, is regulated according to specific cellular requirements through cell-autonomous and non-autonomous signals, and directs transcriptional programs distinct from the HSR during development and in carcinogenesis. We discuss here these 'non-canonical' roles of HSF1, its regulation in diverse conditions of development, reproduction, metabolism, and aging, and posit that HSF1 serves to integrate diverse biological and pathological responses.
Keywords: HSF1; cell proliferation; heat shock response (HSR); metabolism; organismal health; proteostasis.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures


References
-
- Cohen E, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

