Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development
- PMID: 28890333
- PMCID: PMC5610110
- DOI: 10.1016/j.molcel.2017.08.006
Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development
Abstract
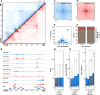
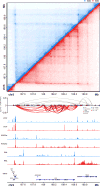
The three-dimensional arrangement of the human genome comprises a complex network of structural and regulatory chromatin loops important for coordinating changes in transcription during human development. To better understand the mechanisms underlying context-specific 3D chromatin structure and transcription during cellular differentiation, we generated comprehensive in situ Hi-C maps of DNA loops in human monocytes and differentiated macrophages. We demonstrate that dynamic looping events are regulatory rather than structural in nature and uncover widespread coordination of dynamic enhancer activity at preformed and acquired DNA loops. Enhancer-bound loop formation and enhancer activation of preformed loops together form multi-loop activation hubs at key macrophage genes. Activation hubs connect 3.4 enhancers per promoter and exhibit a strong enrichment for activator protein 1 (AP-1)-binding events, suggesting that multi-loop activation hubs involving cell-type-specific transcription factors represent an important class of regulatory chromatin structures for the spatiotemporal control of transcription.
Keywords: AP-1; CTCF; DNA looping; Hi-C; cellular differentiation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

