OncoKB: A Precision Oncology Knowledge Base
- PMID: 28890946
- PMCID: PMC5586540
- DOI: 10.1200/PO.17.00011
OncoKB: A Precision Oncology Knowledge Base
Abstract
Purpose: With prospective clinical sequencing of tumors emerging as a mainstay in cancer care, there is an urgent need for a clinical support tool that distills the clinical implications associated with specific mutation events into a standardized and easily interpretable format. To this end, we developed OncoKB, an expert-guided precision oncology knowledge base.
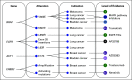
Methods: OncoKB annotates the biological and oncogenic effect and the prognostic and predictive significance of somatic molecular alterations. Potential treatment implications are stratified by the level of evidence that a specific molecular alteration is predictive of drug response based on US Food and Drug Administration (FDA) labeling, National Comprehensive Cancer Network (NCCN) guidelines, disease-focused expert group recommendations and the scientific literature.
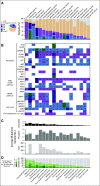
Results: To date, over 3000 unique mutations, fusions, and copy number alterations in 418 cancer-associated genes have been annotated. To test the utility of OncoKB, we annotated all genomic events in 5983 primary tumor samples in 19 cancer types. Forty-one percent of samples harbored at least one potentially actionable alteration, of which 7.5% were predictive of clinical benefit from a standard treatment. OncoKB annotations are available through a public web resource (http://oncokb.org/) and are also incorporated into the cBioPortal for Cancer Genomics to facilitate the interpretation of genomic alterations by physicians and researchers.
Conclusion: OncoKB, a comprehensive and curated precision oncology knowledge base, offers oncologists detailed, evidence-based information about individual somatic mutations and structural alterations present in patient tumors with the goal of supporting optimal treatment decisions.
Conflict of interest statement
Disclosures: Feras M. Hantash and Andrew Grupe are employees of Quest Diagnostics and have some equity interest in the company. All other authors have no pertinent conflicts for the purposes of this paper.
Figures


References
-
- Vanderbilt-Ingram Cancer Center My cancer genome. https://www.mycancergenome.org
-
- The McDonnell Genome Institute: CIViC: Clinical interpretations of variants in cancer. https://civic.genome.wustl.edu
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

