Dockground: A comprehensive data resource for modeling of protein complexes
- PMID: 28891124
- PMCID: PMC5734278
- DOI: 10.1002/pro.3295
Dockground: A comprehensive data resource for modeling of protein complexes
Abstract
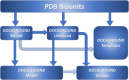
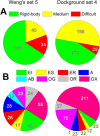
Characterization of life processes at the molecular level requires structural details of protein interactions. The number of experimentally determined structures of protein-protein complexes accounts only for a fraction of known protein interactions. This gap in structural description of the interactome has to be bridged by modeling. An essential part of the development of structural modeling/docking techniques for protein interactions is databases of protein-protein complexes. They are necessary for studying protein interfaces, providing a knowledge base for docking algorithms, and developing intermolecular potentials, search procedures, and scoring functions. Development of protein-protein docking techniques requires thorough benchmarking of different parts of the docking protocols on carefully curated sets of protein-protein complexes. We present a comprehensive description of the Dockground resource (http://dockground.compbio.ku.edu) for structural modeling of protein interactions, including previously unpublished unbound docking benchmark set 4, and the X-ray docking decoy set 2. The resource offers a variety of interconnected datasets of protein-protein complexes and other data for the development and testing of different aspects of protein docking methodologies. Based on protein-protein complexes extracted from the PDB biounit files, Dockground offers sets of X-ray unbound, simulated unbound, model, and docking decoy structures. All datasets are freely available for download, as a whole or selecting specific structures, through a user-friendly interface on one integrated website.
Keywords: benchmark sets; protein recognition; protein-protein interactions; structure prediction.
© 2017 The Protein Society.
Figures




References
-
- Mosca R, Pons T, Ceol A, Valencia A, Aloy P (2013) Towards a detailed atlas of protein–protein interactions. Curr Opin Struct Biol 23:929–940. - PubMed
-
- Moal IH, Moretti R, Baker D, Fernandez‐Recio J (2013) Scoring functions for protein–protein interactions. Curr Opin Struct Biol 23:862–867. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

