No evidence of HIV replication in children on antiretroviral therapy
- PMID: 28891813
- PMCID: PMC5617669
- DOI: 10.1172/JCI94582
No evidence of HIV replication in children on antiretroviral therapy
Abstract
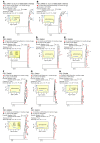
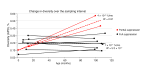
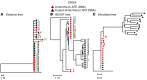
It remains controversial whether current antiretroviral therapy (ART) fully suppresses the cycles of HIV replication and viral evolution in vivo. If replication persists in sanctuary sites such as the lymph nodes, a high priority should be placed on improving ART regimes to target these sites. To investigate the question of ongoing viral replication on current ART regimens, we analyzed HIV populations in longitudinal samples from 10 HIV-1-infected children who initiated ART when viral diversity was low. Eight children started ART at less than ten months of age and showed suppression of plasma viremia for seven to nine years. Two children had uncontrolled viremia for fifteen and thirty months, respectively, before viremia suppression, and served as positive controls for HIV replication and evolution. These latter 2 children showed clear evidence of virus evolution, whereas multiple methods of analysis bore no evidence of virus evolution in any of the 8 children with viremia suppression on ART. Phylogenetic trees simulated with the recently reported evolutionary rate of HIV-1 on ART of 6 × 10-4 substitutions/site/month bore no resemblance to the observed data. Taken together, these data refute the concept that ongoing HIV replication is common with ART and is the major barrier to curing HIV-1 infection.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

