Biodiversity of Actinobacteria from the South Pacific and the Assessment of Streptomyces Chemical Diversity with Metabolic Profiling
- PMID: 28892017
- PMCID: PMC5618425
- DOI: 10.3390/md15090286
Biodiversity of Actinobacteria from the South Pacific and the Assessment of Streptomyces Chemical Diversity with Metabolic Profiling
Abstract
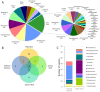
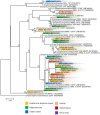
Recently, bioprospecting in underexplored habitats has gained enhanced focus, since new taxa of marine actinobacteria can be found, and thus possible new metabolites. Actinobacteria are in the foreground due to their versatile production of secondary metabolites that present various biological activities, such as antibacterials, antitumorals and antifungals. Chilean marine ecosystems remain largely unexplored and may represent an important source for the discovery of bioactive compounds. Various culture conditions to enrich the growth of this phylum were used and 232 bacterial strains were isolated. Comparative analysis of the 16S rRNA gene sequences led to identifying genetic affiliations of 32 genera, belonging to 20 families. This study shows a remarkable culturable diversity of actinobacteria, associated to marine environments along Chile. Furthermore, 30 streptomycete strains were studied to establish their antibacterial activities against five model strains, Staphylococcus aureus, Listeria monocytogenes, Salmonella enterica, Escherichia coli and Pseudomonas aeruginosa, demonstrating abilities to inhibit bacterial growth of Gram-positive bacteria. To gain insight into their metabolic profiles, crude extracts were submitted to liquid chromatography-high resolution mass spectrometry (LC-HRMS) analysis to assess the selection of streptomycete strains with potentials of producing novel bioactive metabolites. The combined approach allowed for the identification of three streptomycete strains to pursue further investigations. Our Chilean marine actinobacterial culture collection represents an important resource for the bioprospection of novel marine actinomycetes and its metabolites, evidencing their potential as producers of natural bioproducts.
Keywords: Chilean marine actinobacteria; antimicrobial activity; chemical diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Pharmacological Potential of Phylogenetically Diverse Actinobacteria Isolated from Deep-Sea Coral Ecosystems of the Submarine Avilés Canyon in the Cantabrian Sea.Microb Ecol. 2017 Feb;73(2):338-352. doi: 10.1007/s00248-016-0845-2. Epub 2016 Sep 10. Microb Ecol. 2017. PMID: 27614749
-
Bioprospecting of Novel and Bioactive Compounds from Marine Actinomycetes Isolated from South China Sea Sediments.Curr Microbiol. 2018 Feb;75(2):142-149. doi: 10.1007/s00284-017-1358-z. Epub 2017 Sep 16. Curr Microbiol. 2018. PMID: 28918535
-
Antimicrobial potentiality of actinobacteria isolated from two microbiologically unexplored forest ecosystems of Northeast India.BMC Microbiol. 2018 Jul 11;18(1):71. doi: 10.1186/s12866-018-1215-7. BMC Microbiol. 2018. PMID: 29996765 Free PMC article.
-
Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery.Appl Microbiol Biotechnol. 2013 Nov;97(21):9291-321. doi: 10.1007/s00253-013-5229-7. Epub 2013 Sep 22. Appl Microbiol Biotechnol. 2013. PMID: 24057404 Review.
-
Rare taxa and dark microbial matter: novel bioactive actinobacteria abound in Atacama Desert soils.Antonie Van Leeuwenhoek. 2018 Aug;111(8):1315-1332. doi: 10.1007/s10482-018-1088-7. Epub 2018 May 2. Antonie Van Leeuwenhoek. 2018. PMID: 29721711 Review.
Cited by
-
An Integrative Bioinformatic Analysis for Keratinase Detection in Marine-Derived Streptomyces.Mar Drugs. 2021 May 21;19(6):286. doi: 10.3390/md19060286. Mar Drugs. 2021. PMID: 34063876 Free PMC article.
-
Marine Actinobacteria a New Source of Antibacterial Metabolites to Treat Acne Vulgaris Disease-A Systematic Literature Review.Antibiotics (Basel). 2022 Jul 18;11(7):965. doi: 10.3390/antibiotics11070965. Antibiotics (Basel). 2022. PMID: 35884220 Free PMC article. Review.
-
Multiple strain analysis of Streptomyces species from Philippine marine sediments reveals intraspecies heterogeneity in antibiotic activities.Sci Rep. 2021 Sep 2;11(1):17544. doi: 10.1038/s41598-021-96886-4. Sci Rep. 2021. PMID: 34475427 Free PMC article.
-
Genomic Variation and Arsenic Tolerance Emerged as Niche Specific Adaptations by Different Exiguobacterium Strains Isolated From the Extreme Salar de Huasco Environment in Chilean - Altiplano.Front Microbiol. 2020 Jul 15;11:1632. doi: 10.3389/fmicb.2020.01632. eCollection 2020. Front Microbiol. 2020. PMID: 32760381 Free PMC article.
-
Mimicking Nonribosomal Peptides from the Marine Actinomycete Streptomyces sp. H-KF8 Leads to Antimicrobial Peptides.ACS Infect Dis. 2024 Jan 12;10(1):79-92. doi: 10.1021/acsinfecdis.3c00206. Epub 2023 Dec 19. ACS Infect Dis. 2024. PMID: 38113038 Free PMC article.
References
-
- World Health Organization (WHO) Antimicrobial Resistance. Volume 61 World Health Organization; Geneva, Switzerland: 2014.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

