Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection
- PMID: 28892061
- PMCID: PMC6133304
- DOI: 10.1038/ng.3954
Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection
Erratum in
-
Author Correction: Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection.Nat Genet. 2019 Aug;51(8):1295. doi: 10.1038/s41588-019-0468-x. Nat Genet. 2019. PMID: 31273336
Abstract
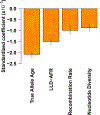
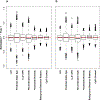
Recent work has hinted at the linkage disequilibrium (LD)-dependent architecture of human complex traits, where SNPs with low levels of LD (LLD) have larger per-SNP heritability. Here we analyzed summary statistics from 56 complex traits (average N = 101,401) by extending stratified LD score regression to continuous annotations. We determined that SNPs with low LLD have significantly larger per-SNP heritability and that roughly half of this effect can be explained by functional annotations negatively correlated with LLD, such as DNase I hypersensitivity sites (DHSs). The remaining signal is largely driven by our finding that more recent common variants tend to have lower LLD and to explain more heritability (P = 2.38 × 10-104); the youngest 20% of common SNPs explain 3.9 times more heritability than the oldest 20%, consistent with the action of negative selection. We also inferred jointly significant effects of other LD-related annotations and confirmed via forward simulations that they jointly predict deleterious effects.
Conflict of interest statement
Competing Financial Interests
N.A.F. is an employee of 23andMe Inc. (Mountain View, CA).
Figures

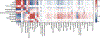

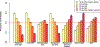
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

