Self-fertility in Chromocrea spinulosa is a consequence of direct repeat-mediated loss of MAT1-2, subsequent imbalance of nuclei differing in mating type, and recognition between unlike nuclei in a common cytoplasm
- PMID: 28892488
- PMCID: PMC5608430
- DOI: 10.1371/journal.pgen.1006981
Self-fertility in Chromocrea spinulosa is a consequence of direct repeat-mediated loss of MAT1-2, subsequent imbalance of nuclei differing in mating type, and recognition between unlike nuclei in a common cytoplasm
Abstract
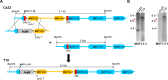
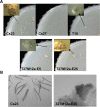
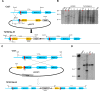
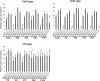
The filamentous fungus Chromocrea spinulosa (Trichoderma spinulosum) exhibits both self-fertile (homothallic) and self-sterile (heterothallic) sexual reproductive behavior. Self-fertile strains produce progeny cohorts that are 50% homothallic, 50% heterothallic. Heterothallic progeny can mate only with homothallic strains, and progeny also segregate 50% homothallic, 50% heterothallic. Sequencing of the mating type (MAT) region of homothallic and heterothallic strains revealed that both carry an intact MAT1-1 locus with three MAT1-1 genes (MAT1-1-1, MAT1-1-2, MAT1-1-3), as previously described for the Sordariomycete group of filamentous fungi. Homothallic strains, however, have a second version of MAT with the MAT1-2 locus genetically linked to MAT1-1. In this version, the MAT1-1-1 open reading frame is split into a large and small fragment and the truncated ends are bordered by 115bp direct repeats (DR). The MAT1-2-1 gene and additional sequences are inserted between the repeats. To understand the mechanism whereby C. spinulosa can exhibit both homothallic and heterothallic behavior, we utilized molecular manipulation to delete one of the DRs from a homothallic strain and insert MAT1-2 into a heterothallic strain. Mating assays indicated that: i) the DRs are key to homothallic behavior, ii) looping out of MAT1-2-1 via intra-molecular homologous recombination between the DRs in self-fertile strains results in two nuclear types in an individual (one carrying both MAT1-1 and MAT1-2 and one carrying MAT1-1 only), iii) self-fertility is achieved by inter-nuclear recognition between these two nuclear types before meiosis, iv) the two types of nuclei are in unequal proportion, v) having both an intact MAT1-1-1 and MAT1-2-1 gene in a single nucleus is not sufficient for self-fertility, and vi) the large truncated MAT1-1-1 fragment is expressed. Comparisons with MAT regions of Trichoderma reesei and Trichoderma virens suggest that several crossovers between misaligned parental MAT chromosomes may have led to the MAT architecture of homothallic C. spinulosa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Altering sexual reproductive mode by interspecific exchange of MAT loci.Fungal Genet Biol. 2011 Jul;48(7):714-24. doi: 10.1016/j.fgb.2011.04.006. Epub 2011 Apr 14. Fungal Genet Biol. 2011. PMID: 21514396
-
Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae.Mol Microbiol. 2003 Oct;50(1):145-52. doi: 10.1046/j.1365-2958.2003.03694.x. Mol Microbiol. 2003. PMID: 14507370
-
The heterothallic sugarbeet pathogen Cercospora beticola contains exon fragments of both MAT genes that are homogenized by concerted evolution.Fungal Genet Biol. 2014 Jan;62:43-54. doi: 10.1016/j.fgb.2013.10.011. Epub 2013 Nov 8. Fungal Genet Biol. 2014. PMID: 24216224
-
Sexual variability in Histoplasma capsulatum and its possible distribution: what is going on?Rev Iberoam Micol. 2014 Jan-Mar;31(1):7-10. doi: 10.1016/j.riam.2013.10.002. Epub 2013 Nov 18. Rev Iberoam Micol. 2014. PMID: 24262630 Review.
-
Aspergillus: sex and recombination.Mycopathologia. 2014 Dec;178(5-6):349-62. doi: 10.1007/s11046-014-9795-8. Epub 2014 Aug 14. Mycopathologia. 2014. PMID: 25118872 Review.
Cited by
-
Comparative Genomics Reveals Evolutionary Traits, Mating Strategies, and Pathogenicity-Related Genes Variation of Botryosphaeriaceae.Front Microbiol. 2022 Feb 23;13:800981. doi: 10.3389/fmicb.2022.800981. eCollection 2022. Front Microbiol. 2022. PMID: 35283828 Free PMC article.
-
Evidence of Biparental Mitochondrial Inheritance from Self-Fertile Crosses between Closely Related Species of Ceratocystis.J Fungi (Basel). 2023 Jun 19;9(6):686. doi: 10.3390/jof9060686. J Fungi (Basel). 2023. PMID: 37367622 Free PMC article.
-
Pneumocystis Mating-Type Locus and Sexual Cycle during Infection.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0000921. doi: 10.1128/MMBR.00009-21. Epub 2021 Jun 16. Microbiol Mol Biol Rev. 2021. PMID: 34132101 Free PMC article. Review.
-
Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia.J Fungi (Basel). 2021 Nov 6;7(11):940. doi: 10.3390/jof7110940. J Fungi (Basel). 2021. PMID: 34829227 Free PMC article.
References
-
- Metzenberg RL, Glass NL (1990) Mating type and mating strategies in Neurospora. Bioessays 12: 53–59. doi: 10.1002/bies.950120202 - DOI - PubMed
-
- Martin T, Lu SW, van Tilbeurgh H, Ripoll DR, Dixelius C, et al. (2010) Tracing the origin of the fungal alpha1 domain places its ancestor in the HMG-box superfamily: implication for fungal mating-type evolution. PLoS One 5: e15199 doi: 10.1371/journal.pone.0015199 - DOI - PMC - PubMed
-
- Nelson MA (1996) Mating systems in ascomycetes: a romp in the sac. Trends Genet 12: 69–74. - PubMed
-
- Turgeon BG (1998) Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol 36: 115–137. doi: 10.1146/annurev.phyto.36.1.115 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

