Chemoproteomics-Enabled Covalent Ligand Screening Reveals a Thioredoxin-Caspase 3 Interaction Disruptor That Impairs Breast Cancer Pathogenicity
- PMID: 28892616
- PMCID: PMC6205226
- DOI: 10.1021/acschembio.7b00711
Chemoproteomics-Enabled Covalent Ligand Screening Reveals a Thioredoxin-Caspase 3 Interaction Disruptor That Impairs Breast Cancer Pathogenicity
Abstract
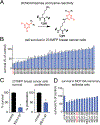
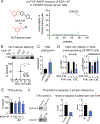
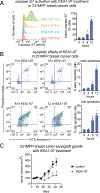
Covalent ligand discovery is a promising strategy to develop small-molecule effectors against therapeutic targets. Recent studies have shown that dichlorotriazines are promising reactive scaffolds that preferentially react with lysines. Here, we have synthesized a series of dichlorotriazine-based covalent ligands and have screened this library to reveal small molecules that impair triple-negative breast cancer cell survival. Upon identifying a lead hit from this screen KEA1-97, we used activity-based protein profiling (ABPP)-based chemoproteomic platforms to identify that this compound targets lysine 72 of thioredoxin-a site previously shown to be important in protein interactions with caspase 3 to inhibit caspase 3 activity and suppress apoptosis. We show that KEA1-97 disrupts the interaction of thioredoxin with caspase 3, activates caspases, and induces apoptosis without affecting thioredoxin activity. Moreover, KEA1-97 impairs in vivo breast tumor xenograft growth. Our study showcases how the screening of covalent ligands can be coupled with ABPP platforms to identify unique anticancer lead and target pairs.
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures
References
-
- Smukste I, and Stockwell BR (2005) Advances in chemical genetics. Annu. Rev. Genomics Hum. Genet. 6, 261–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

