Differentially expressed genes during the imbibition of dormant and after-ripened seeds - a reverse genetics approach
- PMID: 28893189
- PMCID: PMC5594490
- DOI: 10.1186/s12870-017-1098-z
Differentially expressed genes during the imbibition of dormant and after-ripened seeds - a reverse genetics approach
Abstract
Background: Seed dormancy, defined as the incapability of a viable seed to germinate under favourable conditions, is an important trait in nature and agriculture. Despite extensive research on dormancy and germination, many questions about the molecular mechanisms controlling these traits remain unanswered, likely due to its genetic complexity and the large environmental effects which are characteristic of these quantitative traits. To boost research towards revealing mechanisms in the control of seed dormancy and germination we depend on the identification of genes controlling those traits.
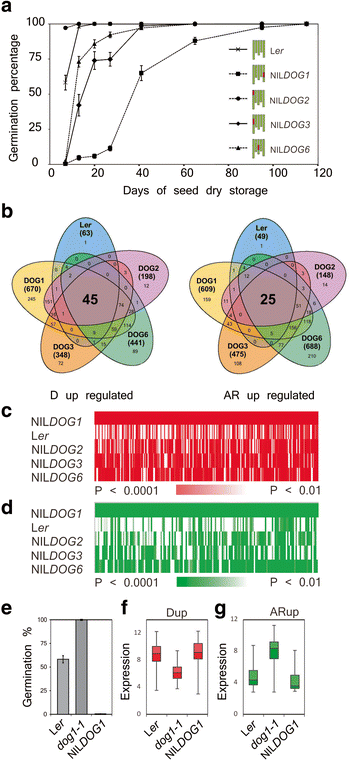
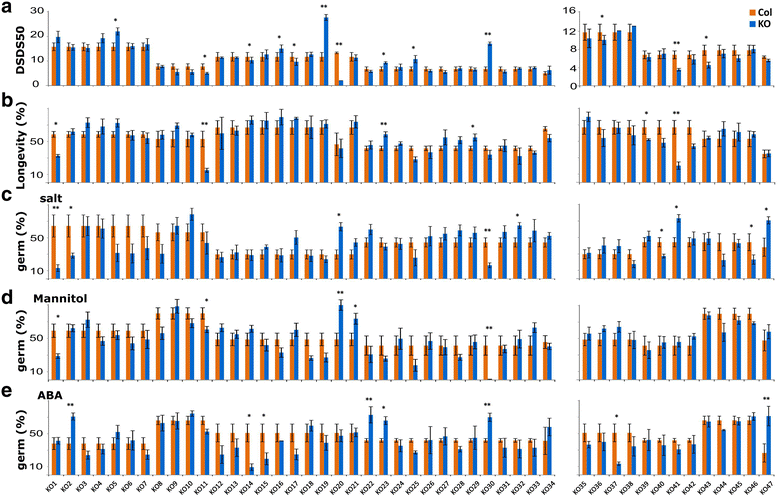
Methods: We used transcriptome analysis combined with a reverse genetics approach to identify genes that are prominent for dormancy maintenance and germination in imbibed seeds of Arabidopsis thaliana. Comparative transcriptomics analysis was employed on freshly harvested (dormant) and after-ripened (AR; non-dormant) 24-h imbibed seeds of four different DELAY OF GERMINATION near isogenic lines (DOGNILs) and the Landsberg erecta (Ler) wild type with varying levels of primary dormancy. T-DNA knock-out lines of the identified genes were phenotypically investigated for their effect on dormancy and AR.
Results: We identified conserved sets of 46 and 25 genes which displayed higher expression in seeds of all dormant and all after-ripened DOGNILs and Ler, respectively. Knock-out mutants in these genes showed dormancy and germination related phenotypes.
Conclusions: Most of the identified genes had not been implicated in seed dormancy or germination. This research will be useful to further decipher the molecular mechanisms by which these important ecological and commercial traits are regulated.
Keywords: Arabidopsis thaliana; Delay of germination; Knockout lines; Seed performance; Transcriptromics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana.Plant J. 2013 Jun;74(5):815-28. doi: 10.1111/tpj.12167. Epub 2013 Apr 4. Plant J. 2013. PMID: 23464703
-
Dormancy cycling: translation-related transcripts are the main difference between dormant and non-dormant seeds in the field.Plant J. 2020 Apr;102(2):327-339. doi: 10.1111/tpj.14626. Epub 2020 Feb 5. Plant J. 2020. PMID: 31785171 Free PMC article.
-
Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds.Plant J. 2010 Feb;61(4):611-22. doi: 10.1111/j.1365-313X.2009.04088.x. Epub 2009 Nov 26. Plant J. 2010. PMID: 19947978
-
Parental and Environmental Control of Seed Dormancy in Arabidopsis thaliana.Annu Rev Plant Biol. 2022 May 20;73:355-378. doi: 10.1146/annurev-arplant-102820-090750. Epub 2022 Feb 9. Annu Rev Plant Biol. 2022. PMID: 35138879 Review.
-
Seed dormancy and germination.Curr Opin Plant Biol. 2002 Feb;5(1):33-6. doi: 10.1016/s1369-5266(01)00219-9. Curr Opin Plant Biol. 2002. PMID: 11788305 Review.
Cited by
-
NADP-Dependent Malic Enzyme 1 Participates in the Abscisic Acid Response in Arabidopsis thaliana.Front Plant Sci. 2018 Nov 6;9:1637. doi: 10.3389/fpls.2018.01637. eCollection 2018. Front Plant Sci. 2018. PMID: 30459802 Free PMC article.
-
An Updated Overview on the Regulation of Seed Germination.Plants (Basel). 2020 Jun 1;9(6):703. doi: 10.3390/plants9060703. Plants (Basel). 2020. PMID: 32492790 Free PMC article. Review.
-
A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination.Plant Cell. 2024 May 1;36(5):1358-1376. doi: 10.1093/plcell/koad328. Plant Cell. 2024. PMID: 38215009 Free PMC article. Review.
-
A New Role for Plastid Thioredoxins in Seed Physiology in Relation to Hormone Regulation.Int J Mol Sci. 2021 Sep 27;22(19):10395. doi: 10.3390/ijms221910395. Int J Mol Sci. 2021. PMID: 34638735 Free PMC article.
-
Late Embryogenesis Abundant Protein-Client Protein Interactions.Plants (Basel). 2020 Jun 29;9(7):814. doi: 10.3390/plants9070814. Plants (Basel). 2020. PMID: 32610443 Free PMC article. Review.
References
-
- Bai B, Novák O, Ljung K, Hanson J, Bentsink L. Combined transcriptome and translatome analyses reveal a role for transcriptional inhibition of tryptophan dependent auxin biosynthesis in the control of DOG1 dependent seed dormancy. 2017. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc B Mec. 1995;57:289–300.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

