Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression
- PMID: 28893265
- PMCID: PMC5594516
- DOI: 10.1186/s12943-017-0719-3
Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression
Abstract
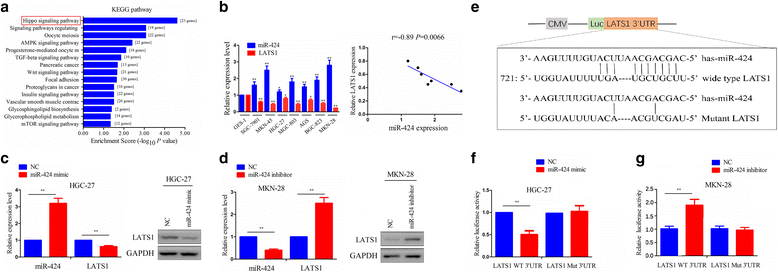
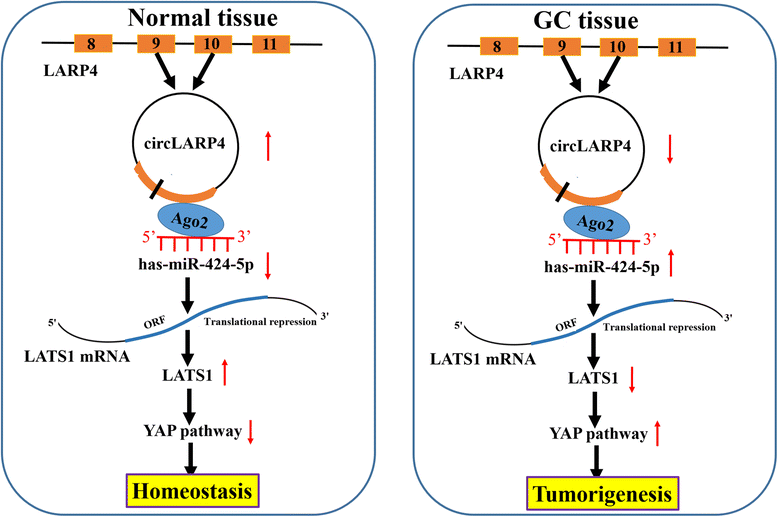
Background: Non-coding RNAs (ncRNAs) have been shown to regulate gene expression involved in tumor progression of multiple malignancies. Our previous studies indicated that large tumor suppressor kinase 1 (LATS1), a core part of Hippo signaling pathway, functions as a tumor suppressor in gastric cancer (GC). But, the underlying molecular mechanisms by which ncRNAs modulate LATS1 expression in GC remain undetermined.
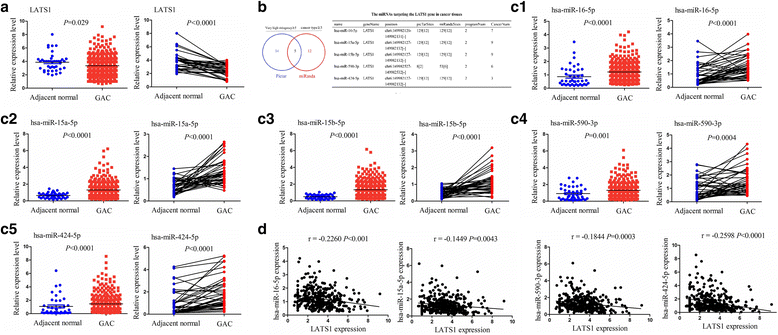
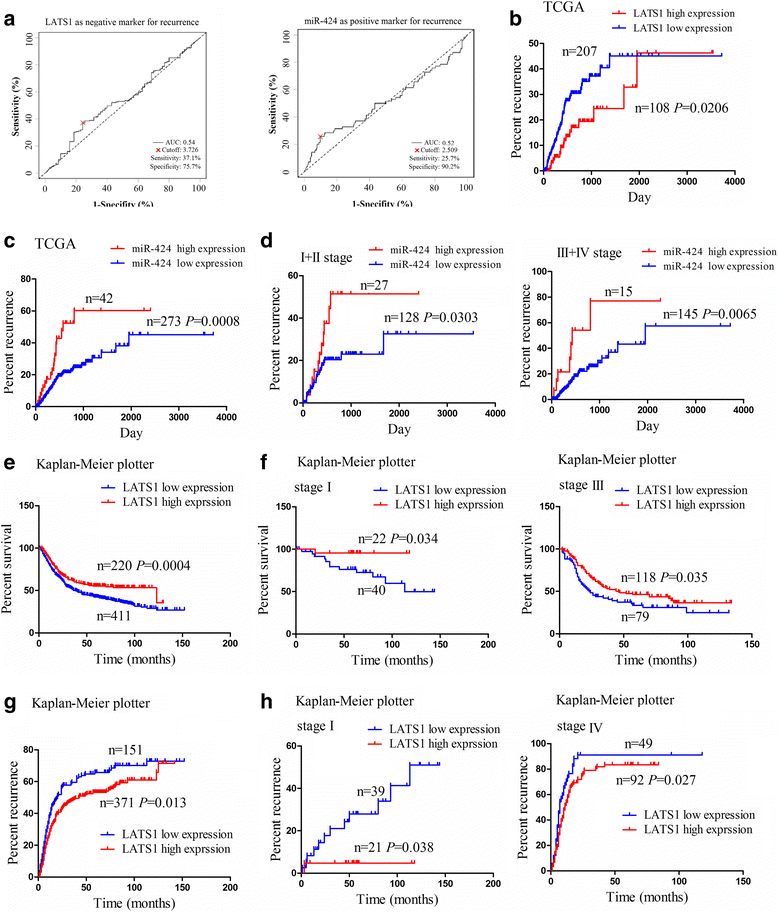
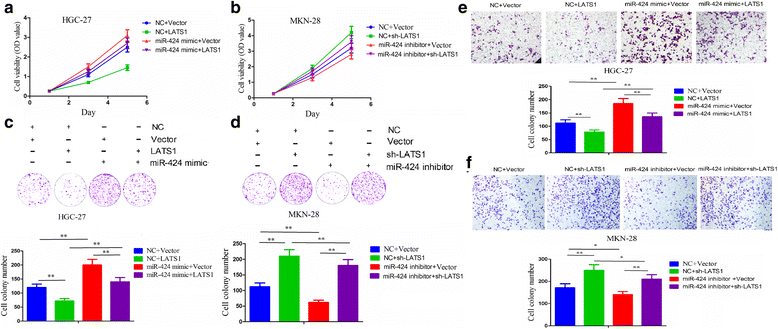
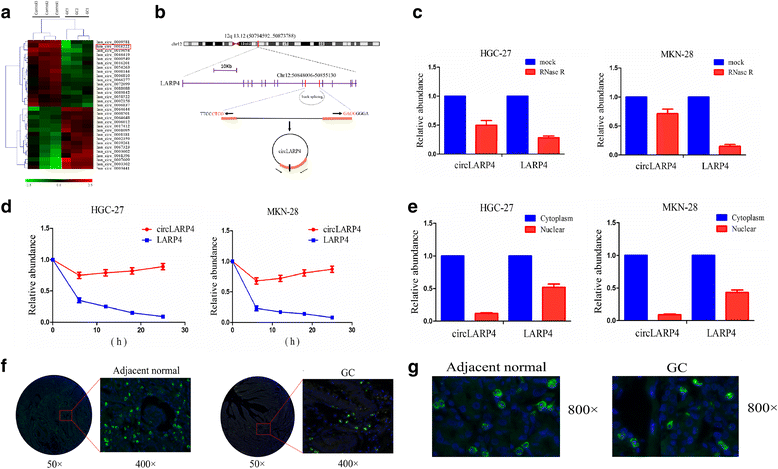
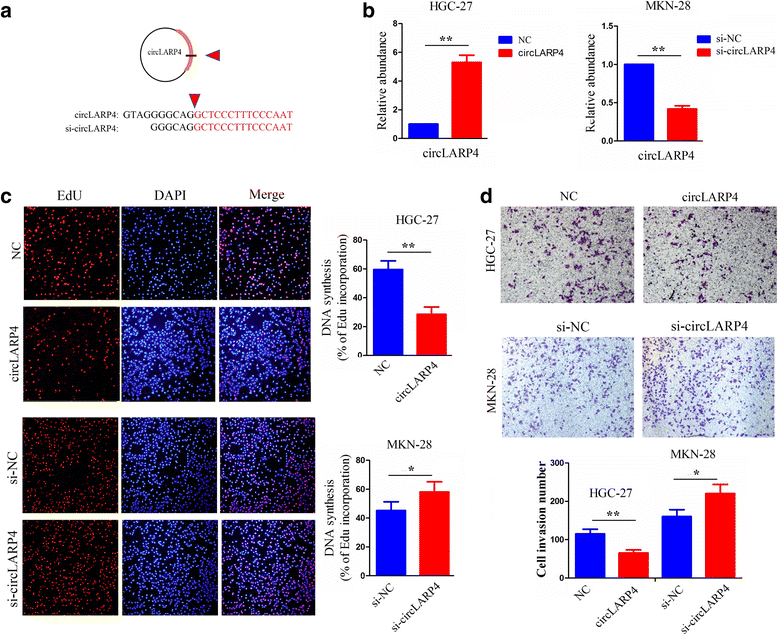
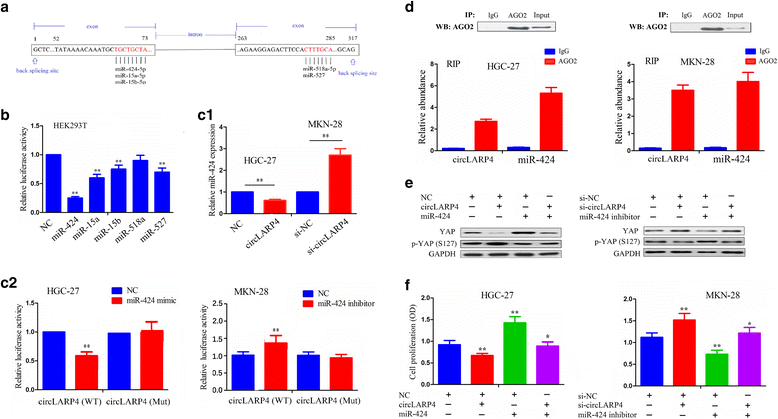
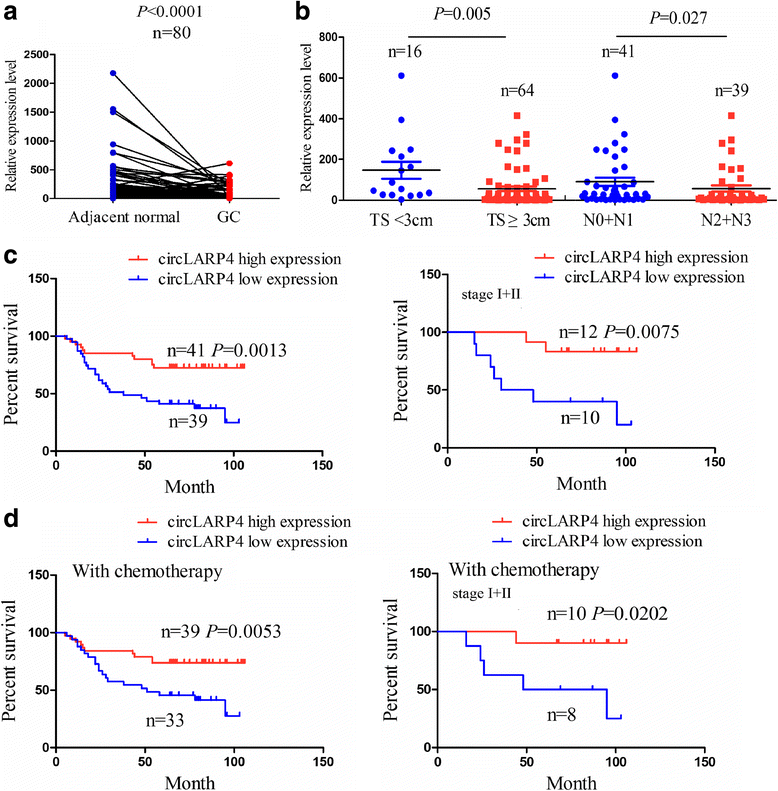
Methods: The correlation of LATS1 and has-miR-424-5p (miR-424) expression with clinicopathological characteristics and prognosis of GC patients was analyzed by TCGA RNA-sequencing data. A novel circular RNA_LARP4 (circLARP4) was identified to sponge miR-424 by circRNA expression profile and bioinformatic analysis. The binding site between miR-424 and LATS1 or circLARP4 was verified using dual luciferase assay and RNA immunoprecipitation (RIP) assay. The expression and localization of circLARP4 in GC tissues were investigated by fluorescence in situ hybridization (FISH). MTT, colony formation, Transwell and EdU assays were performed to assess the effects of miR-424 or circLARP4 on cell proliferation and invasion.
Results: Increased miR-424 expression or decreased LATS1 expression was associated with pathological stage and unfavorable prognosis of GC patients. Ectopic expression of miR-424 promoted proliferation and invasion of GC cells by targeting LATS1 gene. Furthermore, circLARP4 was mainly localized in the cytoplasm and inhibited biological behaviors of GC cells by sponging miR-424. The expression of circLARP4 was downregulated in GC tissues and represented an independent prognostic factor for overall survival of GC patients.
Conclusion: circLARP4 may act as a novel tumor suppressive factor and a potential biomarker in GC.
Keywords: Circular RNA_LARP4; Gastric cancer; Invasion; LATS1; miR-424-5p.
Conflict of interest statement
Ethics approval and consent to participate
The present study was approved by the Hospital’s Protection of Human Subjects Committee.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. Global surveillance of cancer survival 1995e2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385(9972):977–101. doi: 10.1016/S0140-6736(14)62038-9. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

