Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma
- PMID: 28893733
- PMCID: PMC5766840
- DOI: 10.1182/blood-2017-03-770719
Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma
Abstract
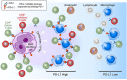
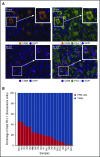
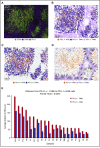
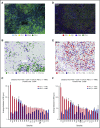
Signaling between programmed cell death protein 1 (PD-1) and the PD-1 ligands (PD-L1, PD-L2) is essential for malignant Hodgkin Reed-Sternberg (HRS) cells to evade antitumor immunity in classical Hodgkin lymphoma (cHL). Copy number alterations of 9p24.1/CD274(PD-L1)/PDCD1LG2(PD-L2) contribute to robust PD-L1 and PD-L2 expression by HRS cells. PD-L1 is also expressed by nonmalignant tumor-associated macrophages (TAMs), but the relationships among PD-L1+ HRS cells, PD-L1+ TAMs, and PD-1+ T cells remain undefined. We used multiplex immunofluorescence and digital image analysis to examine the topography of PD-L1+ and PD-1+ cells in the tumor microenvironment (TME) of cHL. We find that the majority of PD-L1 in the TME is expressed by the abundant PD-L1+ TAMs, which physically colocalize with PD-L1+ HRS cells in a microenvironmental niche. PD-L1+ TAMs are enriched for contacts with T cells, and PD-L1+ HRS cells are enriched for contacts with CD4+ T cells, a subset of which are PD-1+ Our data define a unique topology of cHL in which PD-L1+ TAMs surround HRS cells and implicate CD4+ T cells as a target of PD-1 blockade.
© 2017 by The American Society of Hematology.
Figures

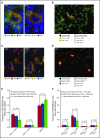
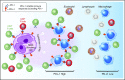
Comment in
-
The ecosystem of classical Hodgkin lymphoma.Blood. 2017 Nov 30;130(22):2360-2361. doi: 10.1182/blood-2017-10-807172. Blood. 2017. PMID: 29191975 No abstract available.
References
-
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2008.
-
- Reichel J, Chadburn A, Rubinstein PG, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood. 2015;125(7):1061-1072. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

