Mitochondrial fission facilitates the selective mitophagy of protein aggregates
- PMID: 28893839
- PMCID: PMC5626535
- DOI: 10.1083/jcb.201612106
Mitochondrial fission facilitates the selective mitophagy of protein aggregates
Abstract
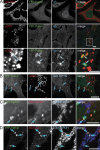
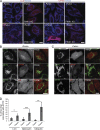
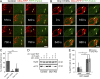
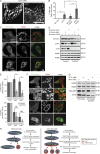
Within the mitochondrial matrix, protein aggregation activates the mitochondrial unfolded protein response and PINK1-Parkin-mediated mitophagy to mitigate proteotoxicity. We explore how autophagy eliminates protein aggregates from within mitochondria and the role of mitochondrial fission in mitophagy. We show that PINK1 recruits Parkin onto mitochondrial subdomains after actinonin-induced mitochondrial proteotoxicity and that PINK1 recruits Parkin proximal to focal misfolded aggregates of the mitochondrial-localized mutant ornithine transcarbamylase (ΔOTC). Parkin colocalizes on polarized mitochondria harboring misfolded proteins in foci with ubiquitin, optineurin, and LC3. Although inhibiting Drp1-mediated mitochondrial fission suppresses the segregation of mitochondrial subdomains containing ΔOTC, it does not decrease the rate of ΔOTC clearance. Instead, loss of Drp1 enhances the recruitment of Parkin to fused mitochondrial networks and the rate of mitophagy as well as decreases the selectivity for ΔOTC during mitophagy. These results are consistent with a new model that, instead of promoting mitophagy, fission protects healthy mitochondrial domains from elimination by unchecked PINK1-Parkin activity.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures






Comment in
-
Splitting up for mitophagy.Nat Cell Biol. 2018 Mar;20(3):224. doi: 10.1038/s41556-018-0058-7. Nat Cell Biol. 2018. PMID: 29476156 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

