Compositional and expression analyses of the glideosome during the Plasmodium life cycle reveal an additional myosin light chain required for maximum motility
- PMID: 28893907
- PMCID: PMC5663884
- DOI: 10.1074/jbc.M117.802769
Compositional and expression analyses of the glideosome during the Plasmodium life cycle reveal an additional myosin light chain required for maximum motility
Abstract
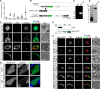
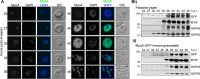
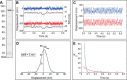
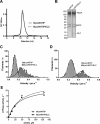
Myosin A (MyoA) is a Class XIV myosin implicated in gliding motility and host cell and tissue invasion by malaria parasites. MyoA is part of a membrane-associated protein complex called the glideosome, which is essential for parasite motility and includes the MyoA light chain myosin tail domain-interacting protein (MTIP) and several glideosome-associated proteins (GAPs). However, most studies of MyoA have focused on single stages of the parasite life cycle. We examined MyoA expression throughout the Plasmodium berghei life cycle in both mammalian and insect hosts. In extracellular ookinetes, sporozoites, and merozoites, MyoA was located at the parasite periphery. In the sexual stages, zygote formation and initial ookinete differentiation precede MyoA synthesis and deposition, which occurred only in the developing protuberance. In developing intracellular asexual blood stages, MyoA was synthesized in mature schizonts and was located at the periphery of segmenting merozoites, where it remained throughout maturation, merozoite egress, and host cell invasion. Besides the known GAPs in the malaria parasite, the complex included GAP40, an additional myosin light chain designated essential light chain (ELC), and several other candidate components. This ELC bound the MyoA neck region adjacent to the MTIP-binding site, and both myosin light chains co-located to the glideosome. Co-expression of MyoA with its two light chains revealed that the presence of both light chains enhances MyoA-dependent actin motility. In conclusion, we have established a system to study the interplay and function of the three glideosome components, enabling the assessment of inhibitors that target this motor complex to block host cell invasion.
Keywords: cell motility; glideosome; invasion; malaria; myosin; myosin light chain; plasmodium.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

