Processive searching ability varies among members of the gap-filling DNA polymerase X family
- PMID: 28893909
- PMCID: PMC5655522
- DOI: 10.1074/jbc.M117.801860
Processive searching ability varies among members of the gap-filling DNA polymerase X family
Abstract
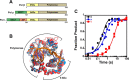
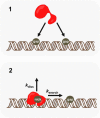
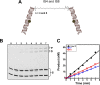
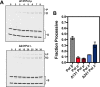
DNA repair proteins must locate rare damaged sites within the genome. DNA polymerase β (Pol β), a member of the DNA polymerase X family that is involved in base excision repair, uses a processive hopping search mechanism to locate substrates. This effectively enhances its search footprint on DNA, increasing the probability of locating damaged sites. Processive searching has been reported or proposed for many DNA-binding proteins, raising the question of how widespread or specific to certain enzymes the ability to perform this function is. To provide insight into this question, we compared the ability of three homologous DNA Pol X family members to perform a processive search for 1-nucleotide gaps in DNA using a previously developed biochemical assay. We found that at near-predicted physiological ionic strengths, the intramolecular searching ability of Pol β is at least 4-fold higher than that of Pol μ and ∼2-fold higher than that of Pol λ. Pol β also was able to perform intersegmental transfer with the intersegmental searching ability of Pol β being at least 6- and ∼2-fold higher than that of Pols μ and λ, respectively. Mutational analysis suggested that differences in the N-terminal domains of these polymerases are responsible for the varying degrees of searching competence. Of note, the differences in processive searching ability observed among the DNA Pol X family members correlated with their proposed biological functions in base excision repair and nonhomologous end joining.
Keywords: DNA binding protein; DNA polymerase; DNA repair; NHEJ; base excision repair (BER); enzyme kinetics; facilitated diffusion; intersegmental transfer; processive search; protein-DNA interaction.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Berg O. G., Winter R. B., and von Hippel P. H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20, 6929–6948 - PubMed
-
- Carey D. C., and Strauss P. R. (1999) Human apurinic/apyrimidinic endonuclease is processive. Biochemistry 38, 16553–16560 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

