In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences
- PMID: 28894115
- PMCID: PMC5593914
- DOI: 10.1038/s41598-017-10724-0
In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences
Abstract
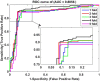
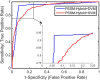
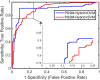
Analysis of drug-target interactions (DTIs) is of great importance in developing new drug candidates for known protein targets or discovering new targets for old drugs. However, the experimental approaches for identifying DTIs are expensive, laborious and challenging. In this study, we report a novel computational method for predicting DTIs using the highly discriminative information of drug-target interactions and our newly developed discriminative vector machine (DVM) classifier. More specifically, each target protein sequence is transformed as the position-specific scoring matrix (PSSM), in which the evolutionary information is retained; then the local binary pattern (LBP) operator is used to calculate the LBP histogram descriptor. For a drug molecule, a novel fingerprint representation is utilized to describe its chemical structure information representing existence of certain functional groups or fragments. When applying the proposed method to the four datasets (Enzyme, GPCR, Ion Channel and Nuclear Receptor) for predicting DTIs, we obtained good average accuracies of 93.16%, 89.37%, 91.73% and 92.22%, respectively. Furthermore, we compared the performance of the proposed model with that of the state-of-the-art SVM model and other previous methods. The achieved results demonstrate that our method is effective and robust and can be taken as a useful tool for predicting DTIs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

