Insm1a Regulates Motor Neuron Development in Zebrafish
- PMID: 28894416
- PMCID: PMC5581358
- DOI: 10.3389/fnmol.2017.00274
Insm1a Regulates Motor Neuron Development in Zebrafish
Abstract
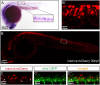
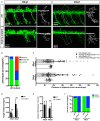
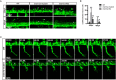
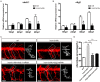
Insulinoma-associated1a (insm1a) is a zinc-finger transcription factor playing a series of functions in cell formation and differentiation of vertebrate central and peripheral nervous systems and neuroendocrine system. However, its roles on the development of motor neuron have still remained uncovered. Here, we provided evidences that insm1a was a vital regulator of motor neuron development, and provided a mechanistic understanding of how it contributes to this process. Firstly, we showed the localization of insm1a in spinal cord, and primary motor neurons (PMNs) of zebrafish embryos by in situ hybridization, and imaging analysis of transgenic reporter line Tg(insm1a: mCherry)ntu805 . Then we demonstrated that the deficiency of insm1a in zebrafish larvae lead to the defects of PMNs development, including the reduction of caudal primary motor neurons (CaP), and middle primary motor neurons (MiP), the excessive branching of motor axons, and the disorganized distance between adjacent CaPs. Additionally, knockout of insm1 impaired motor neuron differentiation in the spinal cord. Locomotion analysis showed that swimming activity was significantly reduced in the insm1a-null zebrafish. Furthermore, we showed that the insm1a loss of function significantly decreased the transcript levels of both olig2 and nkx6.1. Microinjection of olig2 and nkx6.1 mRNA rescued the motor neuron defects in insm1a deficient embryos. Taken together, these data indicated that insm1a regulated the motor neuron development, at least in part, through modulation of the expressions of olig2 and nkx6.1.
Keywords: development; differentiation; insm1a; motor neuron; zebrafish.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

