Fungi found in Mediterranean and North Sea sponges: how specific are they?
- PMID: 28894639
- PMCID: PMC5591636
- DOI: 10.7717/peerj.3722
Fungi found in Mediterranean and North Sea sponges: how specific are they?
Abstract
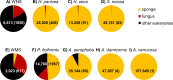
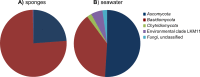
Fungi and other eukaryotes represent one of the last frontiers of microbial diversity in the sponge holobiont. In this study we employed pyrosequencing of 18S ribosomal RNA gene amplicons containing the V7 and V8 hypervariable regions to explore the fungal diversity of seven sponge species from the North Sea and the Mediterranean Sea. For most sponges, fungi were present at a low relative abundance averaging 0.75% of the 18S rRNA gene reads. In total, 44 fungal OTUs (operational taxonomic units) were detected in sponges, and 28 of these OTUs were also found in seawater. Twenty-two of the sponge-associated OTUs were identified as yeasts (mainly Malasseziales), representing 84% of the fungal reads. Several OTUs were related to fungal sequences previously retrieved from other sponges, but all OTUs were also related to fungi from other biological sources, such as seawater, sediments, lakes and anaerobic digesters. Therefore our data, supported by currently available data, point in the direction of mostly accidental presence of fungi in sponges and do not support the existence of a sponge-specific fungal community.
Keywords: Fungi; Malasseziales; Marine sponge; Symbiosis; Yeast.
Conflict of interest statement
Hauke Smidt is an Academic Editor for PeerJ.
Figures



Similar articles
-
Pezizomycotina dominates the fungal communities of South China Sea sponges Theonella swinhoei and Xestospongia testudinaria.FEMS Microbiol Ecol. 2014 Dec;90(3):935-45. doi: 10.1111/1574-6941.12446. Epub 2014 Nov 12. FEMS Microbiol Ecol. 2014. PMID: 25348120
-
Comparisons of the fungal and protistan communities among different marine sponge holobionts by pyrosequencing.Microb Ecol. 2014 May;67(4):951-61. doi: 10.1007/s00248-014-0393-6. Epub 2014 Mar 1. Microb Ecol. 2014. PMID: 24577740
-
Analysis of bacterial communities in sponges and coral inhabiting Red Sea, using barcoded 454 pyrosequencing.Saudi J Biol Sci. 2021 Jan;28(1):847-854. doi: 10.1016/j.sjbs.2020.11.021. Epub 2020 Nov 11. Saudi J Biol Sci. 2021. PMID: 33424375 Free PMC article.
-
Diversity and biotechnological potential of the sponge-associated microbial consortia.J Ind Microbiol Biotechnol. 2006 Jul;33(7):545-51. doi: 10.1007/s10295-006-0123-2. Epub 2006 Apr 22. J Ind Microbiol Biotechnol. 2006. PMID: 16761166 Review.
-
Marine sponge microbial association: Towards disclosing unique symbiotic interactions.Mar Environ Res. 2018 Sep;140:169-179. doi: 10.1016/j.marenvres.2018.04.017. Epub 2018 Apr 27. Mar Environ Res. 2018. PMID: 29935729 Review.
Cited by
-
Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process.Microorganisms. 2020 Aug 20;8(9):1262. doi: 10.3390/microorganisms8091262. Microorganisms. 2020. PMID: 32825344 Free PMC article.
-
First continuous marine sponge cell line established.Sci Rep. 2023 Apr 8;13(1):5766. doi: 10.1038/s41598-023-32394-x. Sci Rep. 2023. PMID: 37031251 Free PMC article.
-
From friends to foes: fungi could be emerging marine sponge pathogens under global change scenarios.Front Microbiol. 2023 Aug 15;14:1213340. doi: 10.3389/fmicb.2023.1213340. eCollection 2023. Front Microbiol. 2023. PMID: 37670990 Free PMC article.
-
Cultivable yeasts associated with marine sponges in the Gulf of Thailand, South China Sea.Antonie Van Leeuwenhoek. 2021 Mar;114(3):253-274. doi: 10.1007/s10482-021-01518-6. Epub 2021 Feb 11. Antonie Van Leeuwenhoek. 2021. PMID: 33575960
-
Metabarcoding assessment of fungal diversity in brown algae and sponges of Mauritius.Front Microbiol. 2022 Oct 28;13:1003790. doi: 10.3389/fmicb.2022.1003790. eCollection 2022. Front Microbiol. 2022. PMID: 36386692 Free PMC article.
References
-
- Borchiellini C, Boury-Esnault N, Vacelet J, Le Parco Y. Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of Metazoa and specific phylogenetic relationships between animals and fungi. Molecular Biology and Evolution. 1998;15:647–655. doi: 10.1093/oxfordjournals.molbev.a025968. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

