Incorporation of Phosphorylated Tyrosine into Proteins: In Vitro Translation and Study of Phosphorylated IκB-α and Its Interaction with NF-κB
- PMID: 28898075
- PMCID: PMC5901656
- DOI: 10.1021/jacs.7b05168
Incorporation of Phosphorylated Tyrosine into Proteins: In Vitro Translation and Study of Phosphorylated IκB-α and Its Interaction with NF-κB
Abstract
Phosphorylated proteins play important roles in the regulation of many different cell networks. However, unlike the preparation of proteins containing unmodified proteinogenic amino acids, which can be altered readily by site-directed mutagenesis and expressed in vitro and in vivo, the preparation of proteins phosphorylated at predetermined sites cannot be done easily and in acceptable yields. To enable the synthesis of phosphorylated proteins for in vitro studies, we have explored the use of phosphorylated amino acids in which the phosphate moiety bears a chemical protecting group, thus eliminating the negative charges that have been shown to have a negative effect on protein translation. Bis-o-nitrobenzyl protection of tyrosine phosphate enabled its incorporation into DHFR and IκB-α using wild-type ribosomes, and the elaborated proteins could subsequently be deprotected by photolysis. Also investigated in parallel was the re-engineering of the 23S rRNA of Escherichia coli, guided by the use of a phosphorylated puromycin, to identify modified ribosomes capable of incorporating unprotected phosphotyrosine into proteins from a phosphotyrosyl-tRNACUA by UAG codon suppression during in vitro translation. Selection of a library of modified ribosomal clones with phosphorylated puromycin identified six modified ribosome variants having mutations in nucleotides 2600-2605 of 23S rRNA; these had enhanced sensitivity to the phosphorylated puromycin. The six clones demonstrated some sequence homology in the region 2600-2605 and incorporated unprotected phosphotyrosine into IκB-α using a modified gene having a TAG codon in the position corresponding to amino acid 42 of the protein. The purified phosphorylated protein bound to a phosphotyrosine specific antibody and permitted NF-κB binding to a DNA duplex sequence corresponding to its binding site in the IL-2 gene promoter. Unexpectedly, phosphorylated IκB-α also mediated the exchange of exogenous DNA into an NF-κB-cellular DNA complex isolated from the nucleus of activated Jurkat cells.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
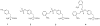



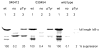








Similar articles
-
Protein Synthesis with Ribosomes Selected for the Incorporation of β-Amino Acids.Biochemistry. 2015 Jun 16;54(23):3694-706. doi: 10.1021/acs.biochem.5b00389. Epub 2015 Jun 2. Biochemistry. 2015. PMID: 25982410 Free PMC article.
-
Expansion of the Genetic Code Through the Use of Modified Bacterial Ribosomes.J Mol Biol. 2022 Apr 30;434(8):167211. doi: 10.1016/j.jmb.2021.167211. Epub 2021 Aug 20. J Mol Biol. 2022. PMID: 34419431 Free PMC article. Review.
-
β-Puromycin selection of modified ribosomes for in vitro incorporation of β-amino acids.Biochemistry. 2012 Jan 10;51(1):401-15. doi: 10.1021/bi2016124. Epub 2011 Dec 19. Biochemistry. 2012. PMID: 22145951
-
23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction.J Bacteriol. 2009 Jun;191(11):3445-50. doi: 10.1128/JB.00096-09. Epub 2009 Mar 27. J Bacteriol. 2009. PMID: 19329641 Free PMC article.
-
Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity.Annu Rev Immunol. 2000;18:621-63. doi: 10.1146/annurev.immunol.18.1.621. Annu Rev Immunol. 2000. PMID: 10837071 Review.
Cited by
-
Genetic Encoding of Phosphorylated Amino Acids into Proteins.Chem Rev. 2024 May 22;124(10):6592-6642. doi: 10.1021/acs.chemrev.4c00110. Epub 2024 May 1. Chem Rev. 2024. PMID: 38691379 Free PMC article. Review.
-
Puromycins B-E, Naturally Occurring Amino-Nucleosides Produced by the Himalayan Isolate Streptomyces sp. PU-14G.J Nat Prod. 2018 Nov 26;81(11):2560-2566. doi: 10.1021/acs.jnatprod.8b00720. Epub 2018 Nov 12. J Nat Prod. 2018. PMID: 30418763 Free PMC article.
-
Deciphering protein post-translational modifications using chemical biology tools.Nat Rev Chem. 2020 Dec;4(12):674-695. doi: 10.1038/s41570-020-00223-8. Epub 2020 Oct 6. Nat Rev Chem. 2020. PMID: 37127974 Review.
-
Elongation Factor P Modulates the Incorporation of Structurally Diverse Noncanonical Amino Acids into Escherichia coli Dihydrofolate Reductase.J Am Chem Soc. 2023 Nov 1;145(43):23600-23608. doi: 10.1021/jacs.3c07524. Epub 2023 Oct 23. J Am Chem Soc. 2023. PMID: 37871253 Free PMC article.
-
Mechanistic studies of non-canonical amino acid mutagenesis.Methods Enzymol. 2021;656:375-428. doi: 10.1016/bs.mie.2021.05.001. Epub 2021 Jun 24. Methods Enzymol. 2021. PMID: 34325793 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

