Transcriptional Regulation by Nrf2
- PMID: 28899199
- PMCID: PMC6208165
- DOI: 10.1089/ars.2017.7342
Transcriptional Regulation by Nrf2
Abstract
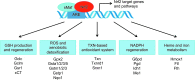
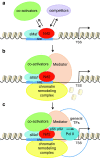
Significance: Nuclear factor E2-related factor 2 (Nrf2) is a transcription factor that coordinates the basal and stress-inducible activation of a vast array of cytoprotective genes. Understanding the regulation of Nrf2 activity and downstream pathways has major implications for human health. Recent Advances: Nrf2 regulates the transcription of components of the glutathione and thioredoxin antioxidant systems, as well as enzymes involved in phase I and phase II detoxification of exogenous and endogenous products, NADPH regeneration, and heme metabolism. It therefore represents a crucial regulator of the cellular defense mechanisms against xenobiotic and oxidative stress. In addition to antioxidant responses, Nrf2 is involved in other cellular processes, such as autophagy, intermediary metabolism, stem cell quiescence, and unfolded protein response. Given the wide range of processes that Nrf2 controls, its activity is tightly regulated at multiple levels. Here, we review the different modes of regulation of Nrf2 activity and the current knowledge of Nrf2-mediated transcriptional control.
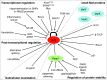
Critical issues: It is now clear that Nrf2 lies at the center of a complex regulatory network. A full comprehension of the Nrf2 program will require an integrated consideration of all the different factors determining Nrf2 activity.
Future directions: Additional computational and experimental studies are needed to obtain a more dynamic global view of Nrf2-mediated gene regulation. In particular, studies comparing how the Nrf2-dependent network changes from a physiological to a pathological condition can provide insight into mechanisms of disease and instruct new treatment strategies.
Keywords: Nrf2; antioxidant response; transcription.
Figures



References
-
- Abbas K, Breton J, Planson AG, Bouton C, Bignon J, Seguin C, Riquier S, Toledano MB, and Drapier JC. Nitric oxide activates an Nrf2/sulfiredoxin antioxidant pathway in macrophages. Free Radic Biol Med 51: 107–114, 2011 - PubMed
-
- Ahmad R, Raina D, Meyer C, Kharbanda S, and Kufe D. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem 281: 35764–35769, 2006 - PubMed
-
- Alam J, Killeen E, Gong P, Naquin R, Hu B, Stewart D, Ingelfinger JR, and Nath KA. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am J Physiol Renal Physiol 284: F743–F752, 2003 - PubMed
-
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, and Cook JL. Nrf2, a Cap ‘n’ Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 274: 26071–26078, 1999 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

