Structure and function of Pif1 helicase
- PMID: 28900015
- PMCID: PMC5870758
- DOI: 10.1042/BST20170096
Structure and function of Pif1 helicase
Abstract
Pif1 family helicases have multiple roles in the maintenance of nuclear and mitochondrial DNA in eukaryotes. Saccharomyces cerevisiae Pif1 is involved in replication through barriers to replication, such as G-quadruplexes and protein blocks, and reduces genetic instability at these sites. Another Pif1 family helicase in S. cerevisiae, Rrm3, assists in fork progression through replication fork barriers at the rDNA locus and tRNA genes. ScPif1 (Saccharomyces cerevisiae Pif1) also negatively regulates telomerase, facilitates Okazaki fragment processing, and acts with polymerase δ in break-induced repair. Recent crystal structures of bacterial Pif1 helicases and the helicase domain of human PIF1 combined with several biochemical and biological studies on the activities of Pif1 helicases have increased our understanding of the function of these proteins. This review article focuses on these structures and the mechanism(s) proposed for Pif1's various activities on DNA.
Keywords: DNA; G-quadruplex; enzyme kinetics; enzyme–substrate interactions; helicase; mtDNA.
© 2017 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
Declarations of interest
The authors declare that conflict of interest associated with this manuscript.
Figures
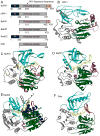
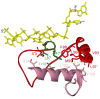
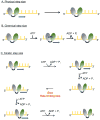
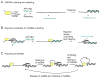
References
-
- Bessler JB, Torredagger JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 2001;11:60–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

