A 3D magnetic tissue stretcher for remote mechanical control of embryonic stem cell differentiation
- PMID: 28900152
- PMCID: PMC5596024
- DOI: 10.1038/s41467-017-00543-2
A 3D magnetic tissue stretcher for remote mechanical control of embryonic stem cell differentiation
Abstract
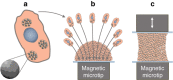
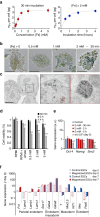
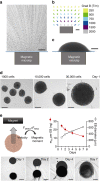
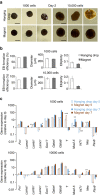
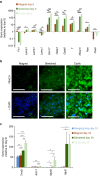
The ability to create a 3D tissue structure from individual cells and then to stimulate it at will is a major goal for both the biophysics and regenerative medicine communities. Here we show an integrated set of magnetic techniques that meet this challenge using embryonic stem cells (ESCs). We assessed the impact of magnetic nanoparticles internalization on ESCs viability, proliferation, pluripotency and differentiation profiles. We developed magnetic attractors capable of aggregating the cells remotely into a 3D embryoid body. This magnetic approach to embryoid body formation has no discernible impact on ESC differentiation pathways, as compared to the hanging drop method. It is also the base of the final magnetic device, composed of opposing magnetic attractors in order to form embryoid bodies in situ, then stretch them, and mechanically stimulate them at will. These stretched and cyclic purely mechanical stimulations were sufficient to drive ESCs differentiation towards the mesodermal cardiac pathway.The development of embryoid bodies that are responsive to external stimuli is of great interest in tissue engineering. Here, the authors culture embryonic stem cells with magnetic nanoparticles and show that the presence of magnetic fields could affect their aggregation and differentiation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

