Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells
- PMID: 28900194
- PMCID: PMC5595809
- DOI: 10.1038/s41598-017-10843-8
Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells
Abstract
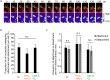
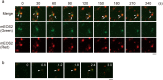
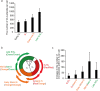
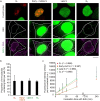
Mitochondrial DNA (mtDNA) is organized in nucleoprotein complexes called mitochondrial nucleoids (mt-nucleoids), which are critical units of mtDNA replication and transmission. In humans, several hundreds of mt-nucleoids exist in a cell. However, how numerous mt-nucleoids are maintained during the cell cycle remains elusive, because cell cycle synchronization procedures affect mtDNA replication. Here, we analyzed regulation of the maintenance of mt-nucleoids in the cell cycle, using a fluorescent cell cycle indicator, Fucci2. Live imaging of mt-nucleoids with higher temporal resolution showed frequent attachment and detachment of mt-nucleoids throughout the cell cycle. TFAM, an mtDNA packaging protein, was involved in the regulation of this dynamic process, which was important for maintaining proper mt-nucleoid number. Both an increase in mt-nucleoid number and activation of mtDNA replication occurred during S phase. To increase mt-nucleoid number, mtDNA replication, but not nuclear DNA replication, was necessary. We propose that these dynamic and regulatory processes in the cell cycle maintain several hundred mt-nucleoids in proliferating cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 2010;51:440–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

