Complete Genome Sequence of Geobacillus thermodenitrificans T12, A Potential Host for Biotechnological Applications
- PMID: 28900693
- PMCID: PMC5765199
- DOI: 10.1007/s00284-017-1349-0
Complete Genome Sequence of Geobacillus thermodenitrificans T12, A Potential Host for Biotechnological Applications
Abstract
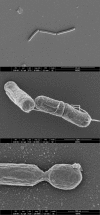
In attempt to obtain a thermophilic host for the conversion of lignocellulose derived substrates into lactic acid, Geobacillus thermodenitrificans T12 was isolated from a compost heap. It was selected from over 500 isolates as a genetically tractable hemicellulolytic lactic acid producer, requiring little nutrients. The strain is able to ferment glucose and xylose simultaneously and can produce lactic acid from xylan, making it a potential host for biotechnological applications. The genome of strain T12 consists of a 3.64 Mb chromosome and two plasmids of 59 and 56 kb. It has a total of 3.676 genes with an average genomic GC content of 48.7%. The T12 genome encodes a denitrification pathway, allowing for anaerobic respiration. The identity and localization of the responsible genes are similar to those of the denitrification pathways found in strain NG80-2. The hemicellulose utilization (HUS) locus was identified based on sequence homology against G. stearothermophilus T-6. It appeared that T12 has all the genes that are present in strain T-6 except for the arabinan degradation cluster. Instead, the HUS locus of strain T12 contains genes for both an inositol and a pectate degradation pathway. Strain T12 has complete pathways for the synthesis of purine and pyrimidine, all 20 amino acids and several vitamins except D-biotin. The host-defense systems present comprise a Type II and a Type III restriction-modification system, as well as a CRISPR-Cas Type II system. It is concluded that G. thermodenitrificans T12 is a potentially interesting candidate for industrial applications.
Conflict of interest statement
Conflict of interest
The authors declare to have no conflict of interest. RvK and BV are employed by the biotech company Corbion (Gorinchem, The Netherlands).
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures



References
-
- Banat IM, Marchant R, Rahman TJ. Geobacillus debilis sp. nov., a novel obligately thermophilic bacterium isolated from a cool soil environment, and reassignment of Bacillus pallidus to Geobacillus pallidus comb. nov. Int J Syst Evol Microbiol. 2004;54:2197–2201. doi: 10.1099/ijs.0.63231-0. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

