The Apostasia genome and the evolution of orchids
- PMID: 28902843
- PMCID: PMC7416622
- DOI: 10.1038/nature23897
The Apostasia genome and the evolution of orchids
Erratum in
-
Author Correction: The Apostasia genome and the evolution of orchids.Nature. 2020 Jul;583(7818):E30. doi: 10.1038/s41586-020-2524-1. Nature. 2020. PMID: 32681116 Free PMC article.
Abstract
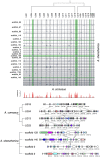
Constituting approximately 10% of flowering plant species, orchids (Orchidaceae) display unique flower morphologies, possess an extraordinary diversity in lifestyle, and have successfully colonized almost every habitat on Earth. Here we report the draft genome sequence of Apostasia shenzhenica, a representative of one of two genera that form a sister lineage to the rest of the Orchidaceae, providing a reference for inferring the genome content and structure of the most recent common ancestor of all extant orchids and improving our understanding of their origins and evolution. In addition, we present transcriptome data for representatives of Vanilloideae, Cypripedioideae and Orchidoideae, and novel third-generation genome data for two species of Epidendroideae, covering all five orchid subfamilies. A. shenzhenica shows clear evidence of a whole-genome duplication, which is shared by all orchids and occurred shortly before their divergence. Comparisons between A. shenzhenica and other orchids and angiosperms also permitted the reconstruction of an ancestral orchid gene toolkit. We identify new gene families, gene family expansions and contractions, and changes within MADS-box gene classes, which control a diverse suite of developmental processes, during orchid evolution. This study sheds new light on the genetic mechanisms underpinning key orchid innovations, including the development of the labellum and gynostemium, pollinia, and seeds without endosperm, as well as the evolution of epiphytism; reveals relationships between the Orchidaceae subfamilies; and helps clarify the evolutionary history of orchids within the angiosperms.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
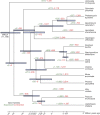
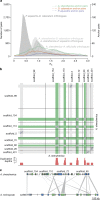
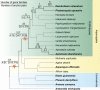
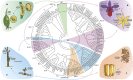


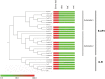

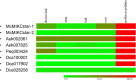

References
-
- Roberts DL, Dixon KW. Orchids. Curr. Biol. 2008;18:R325–R329. - PubMed
-
- Givnish TJ, et al. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr. 2016;43:1905–1916.
-
- Chen LJ, Liu ZJ. Apostasia shenzhenica: a new species of Apostasioideae (Orchidaceae) from China. Plant Science Journal. 2011;29:38–41.
-
- Kocyan A, Qiu Y-L, Endress PK, Conti E. A phylogenetic analysis of Apostasioideae (Orchidaceae) based on ITS, trnL-F and matK sequences. Plant Syst. Evol. 2004;247:203–213.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

