Histone modification alteration coordinated with acquisition of promoter DNA methylation during Epstein-Barr virus infection
- PMID: 28903418
- PMCID: PMC5589657
- DOI: 10.18632/oncotarget.19423
Histone modification alteration coordinated with acquisition of promoter DNA methylation during Epstein-Barr virus infection
Abstract
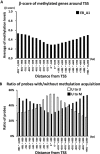
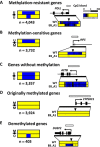
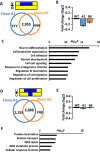
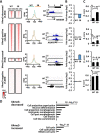
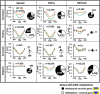
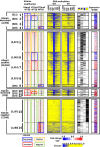
Aberrant DNA hypermethylation is a major epigenetic mechanism to inactivate tumor suppressor genes in cancer. Epstein-Barr virus positive gastric cancer is the most frequently hypermethylated tumor among human malignancies. Herein, we performed comprehensive analysis of epigenomic alteration during EBV infection, by Infinium HumanMethylation 450K BeadChip for DNA methylation and ChIP-sequencing for histone modification alteration during EBV infection into gastric cancer cell line MKN7. Among 7,775 genes with increased DNA methylation in promoter regions, roughly half were "DNA methylation-sensitive" genes, which acquired DNA methylation in the whole promoter regions and thus were repressed. These included anti-oncogenic genes, e.g. CDKN2A. The other half were "DNA methylation-resistant" genes, where DNA methylation is acquired in the surrounding of promoter regions, but unmethylated status is protected in the vicinity of transcription start site. These genes thereby retained gene expression, and included DNA repair genes. Histone modification was altered dynamically and coordinately with DNA methylation alteration. DNA methylation-sensitive genes significantly correlated with loss of H3K27me3 pre-marks or decrease of active histone marks, H3K4me3 and H3K27ac. Apoptosis-related genes were significantly enriched in these epigenetically repressed genes. Gain of active histone marks significantly correlated with DNA methylation-resistant genes. Genes related to mitotic cell cycle and DNA repair were significantly enriched in these epigenetically activated genes. Our data show that orchestrated epigenetic alterations are important in gene regulation during EBV infection, and histone modification status in promoter regions significantly associated with acquisition of de novo DNA methylation or protection of unmethylated status at transcription start site.
Keywords: DNA methylation; Epstein-Barr virus; gastric cancer.
Conflict of interest statement
CONFLICTS OF INTEREST All authors have no potential conflicts of interest to disclose.
Figures


References
-
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. - PubMed
-
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. - PubMed
-
- Funata S, Fukayama M, Kaneda A. De NovoMethylation in. Cancer. 2016;eLS:1–7. https://doi.org/10.1002/9780470015902.a0026465. - DOI
-
- Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H, Fukayama M. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–7197. - PubMed
-
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

