A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome
- PMID: 28903732
- PMCID: PMC5598039
- DOI: 10.1186/s12866-017-1101-8
A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome
Abstract
Background: Advancements in Next Generation Sequencing (NGS) technologies regarding throughput, read length and accuracy had a major impact on microbiome research by significantly improving 16S rRNA amplicon sequencing. As rapid improvements in sequencing platforms and new data analysis pipelines are introduced, it is essential to evaluate their capabilities in specific applications. The aim of this study was to assess whether the same project-specific biological conclusions regarding microbiome composition could be reached using different sequencing platforms and bioinformatics pipelines.
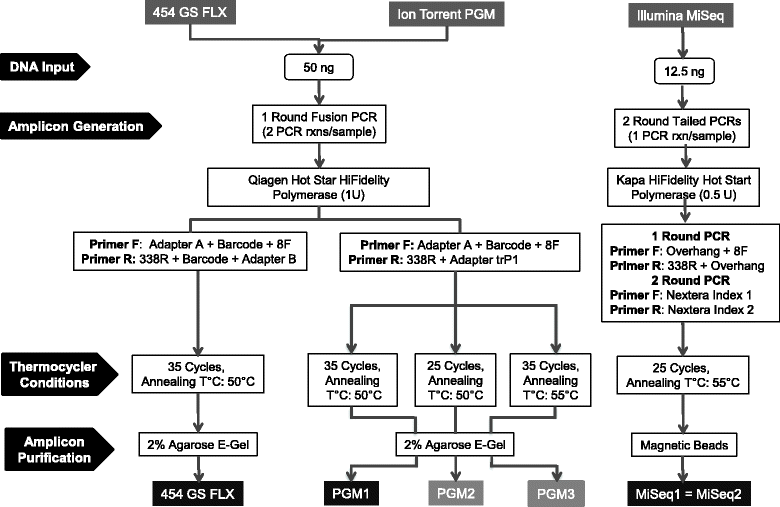
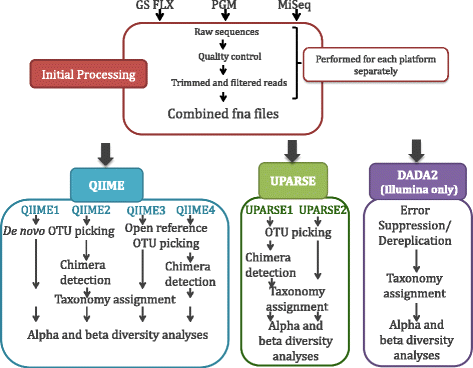
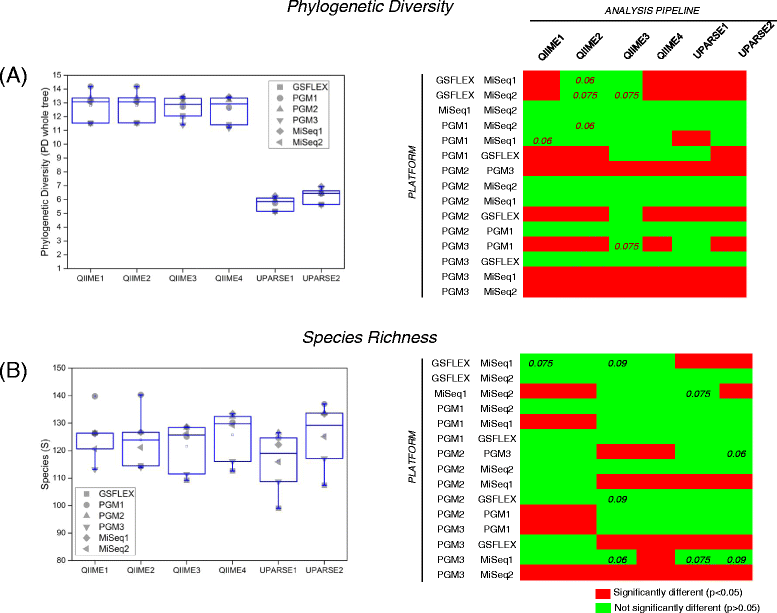
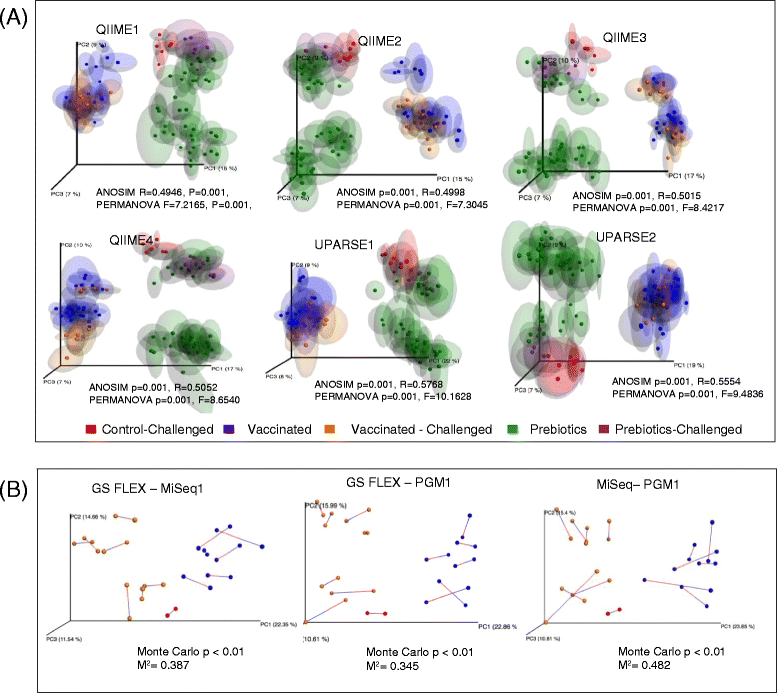
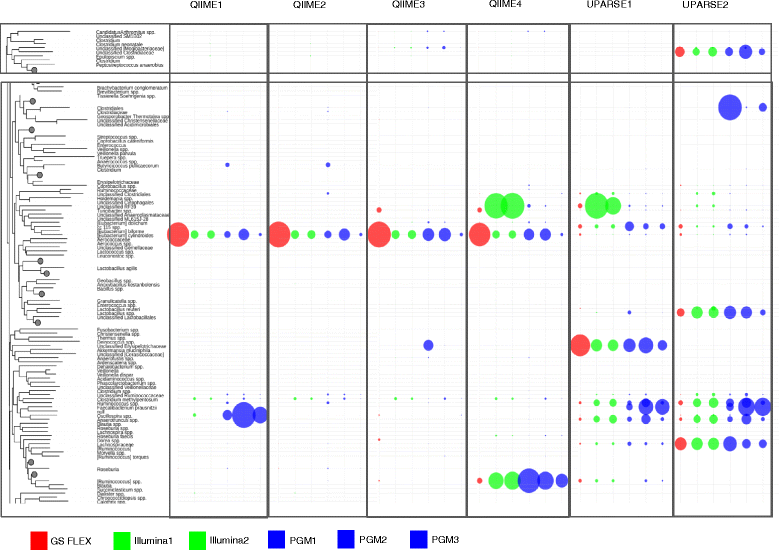
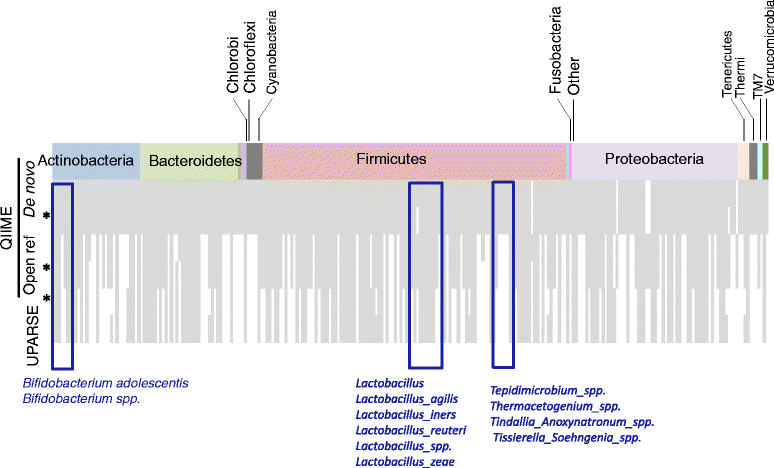
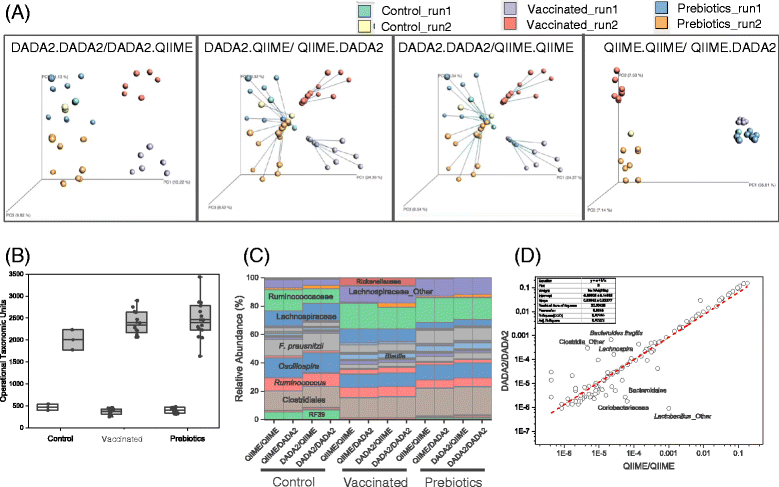
Results: Chicken cecum microbiome was analyzed by 16S rRNA amplicon sequencing using Illumina MiSeq, Ion Torrent PGM, and Roche 454 GS FLX Titanium platforms, with standard and modified protocols for library preparation. We labeled the bioinformatics pipelines included in our analysis QIIME1 and QIIME2 (de novo OTU picking [not to be confused with QIIME version 2 commonly referred to as QIIME2]), QIIME3 and QIIME4 (open reference OTU picking), UPARSE1 and UPARSE2 (each pair differs only in the use of chimera depletion methods), and DADA2 (for Illumina data only). GS FLX+ yielded the longest reads and highest quality scores, while MiSeq generated the largest number of reads after quality filtering. Declines in quality scores were observed starting at bases 150-199 for GS FLX+ and bases 90-99 for MiSeq. Scores were stable for PGM-generated data. Overall microbiome compositional profiles were comparable between platforms; however, average relative abundance of specific taxa varied depending on sequencing platform, library preparation method, and bioinformatics analysis. Specifically, QIIME with de novo OTU picking yielded the highest number of unique species and alpha diversity was reduced with UPARSE and DADA2 compared to QIIME.
Conclusions: The three platforms compared in this study were capable of discriminating samples by treatment, despite differences in diversity and abundance, leading to similar biological conclusions. Our results demonstrate that while there were differences in depth of coverage and phylogenetic diversity, all workflows revealed comparable treatment effects on microbial diversity. To increase reproducibility and reliability and to retain consistency between similar studies, it is important to consider the impact on data quality and relative abundance of taxa when selecting NGS platforms and analysis tools for microbiome studies.
Keywords: 16S rRNA amplicon sequencing - microbiome analysis - microbiome - microbiome composition - next generation sequencing platforms; Bioinformatics pipeline; NGS bias.
Conflict of interest statement
Ethics approval
Animals were euthanized according to protocol #15–065-A, approved by the North Carolina State University Institutional Animal Care and Use Committee (OLAW# D16–00214) and sampled for gut microbiome analysis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. 2015;5:3. doi: 10.3389/fcimb.2015.00003. - DOI - PMC - PubMed
-
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

