Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy
- PMID: 28903782
- PMCID: PMC5598015
- DOI: 10.1186/s13059-017-1286-z
Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy
Abstract
Background: Genetic variation is an important determinant of RNA transcription and splicing, which in turn contributes to variation in human traits, including cardiovascular diseases.
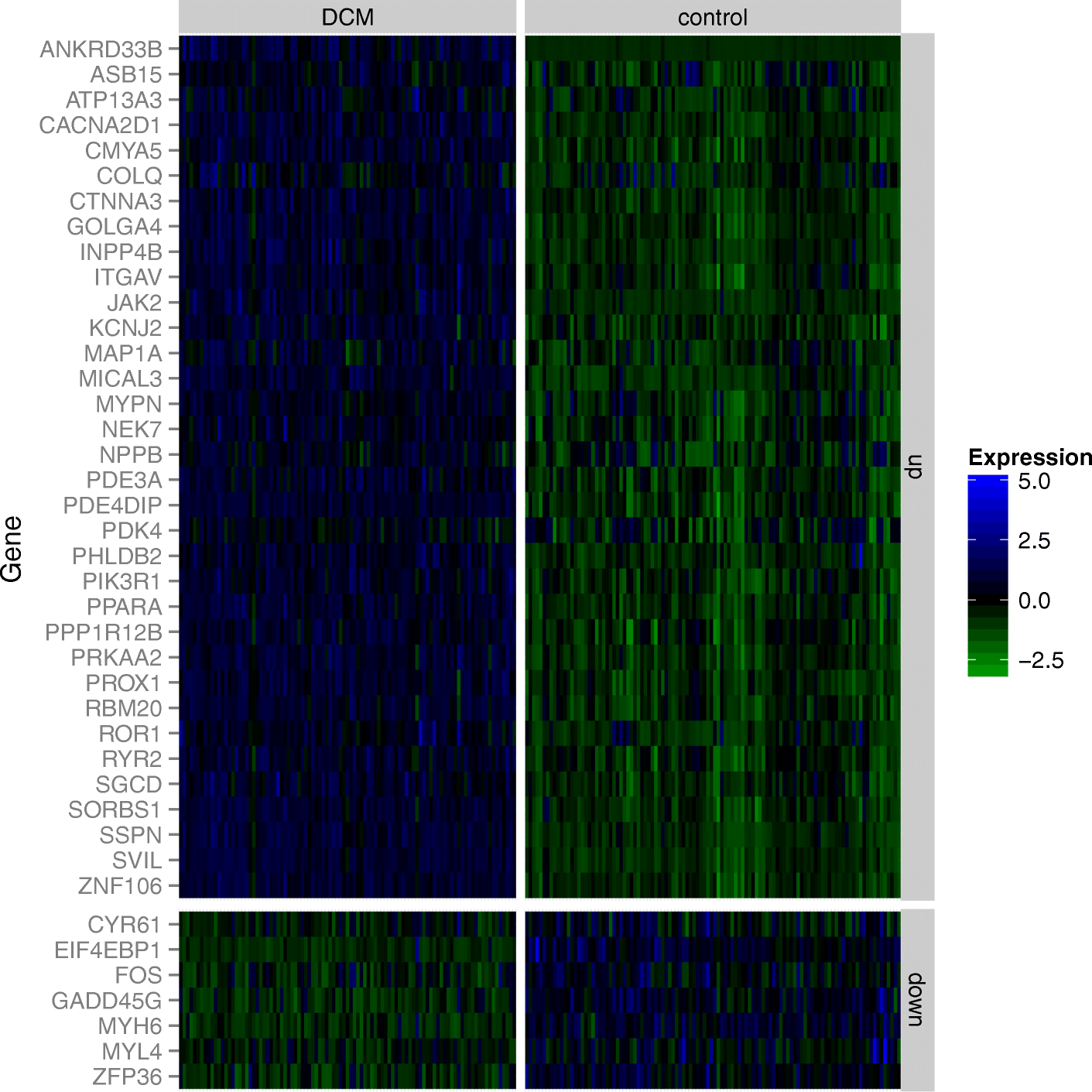
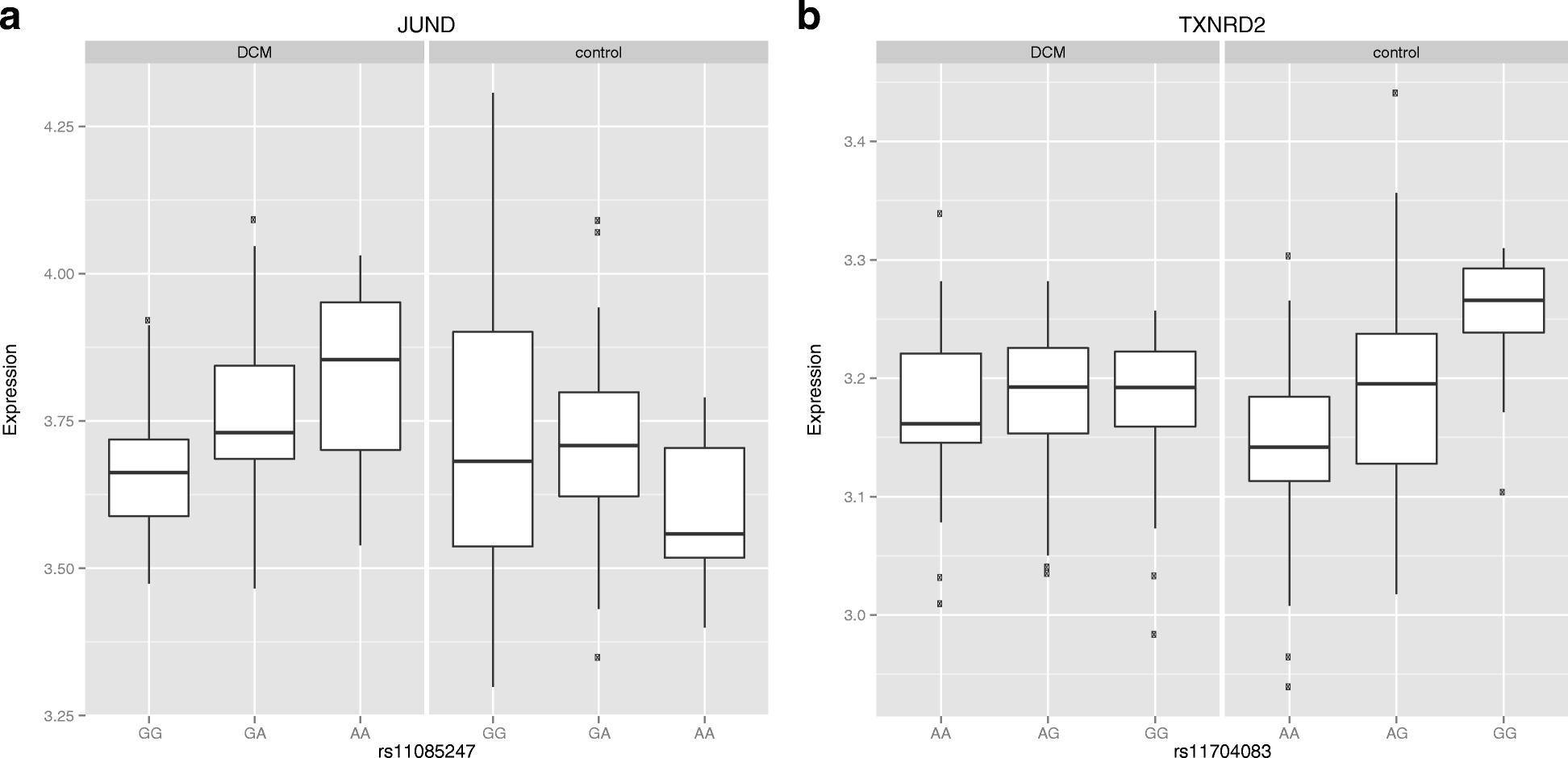
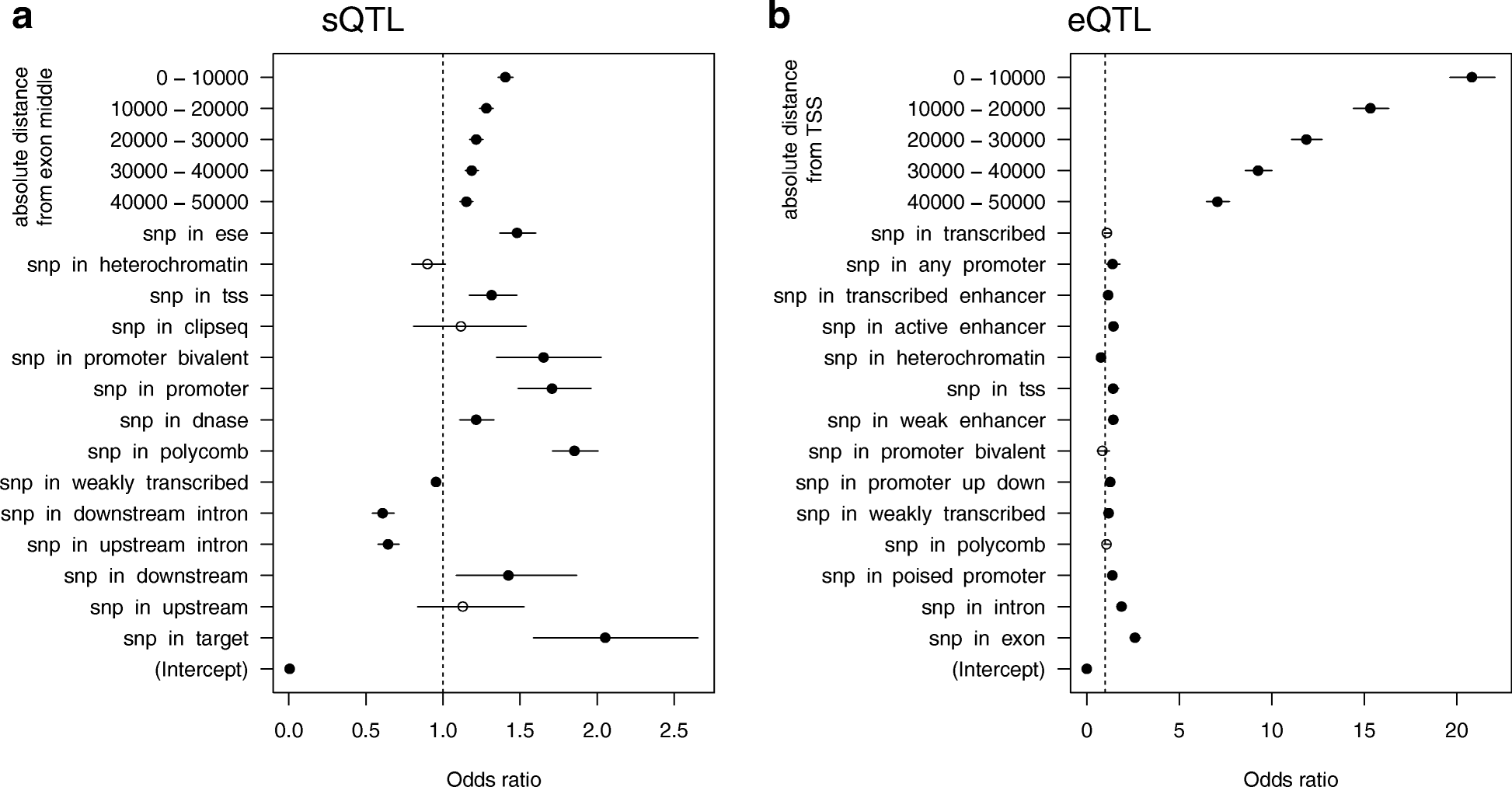
Results: Here we report the first in-depth survey of heart transcriptome variation using RNA-sequencing in 97 patients with dilated cardiomyopathy and 108 non-diseased controls. We reveal extensive differences of gene expression and splicing between dilated cardiomyopathy patients and controls, affecting known as well as novel dilated cardiomyopathy genes. Moreover, we show a widespread effect of genetic variation on the regulation of transcription, isoform usage, and allele-specific expression. Systematic annotation of genome-wide association SNPs identifies 60 functional candidate genes for heart phenotypes, representing 20% of all published heart genome-wide association loci. Focusing on the dilated cardiomyopathy phenotype we found that eQTL variants are also enriched for dilated cardiomyopathy genome-wide association signals in two independent cohorts.
Conclusions: RNA transcription, splicing, and allele-specific expression are each important determinants of the dilated cardiomyopathy phenotype and are controlled by genetic factors. Our results represent a powerful resource for the field of cardiovascular genetics.
Keywords: Dilated cardiomyopathy; Gene expression; Genetics; Heart; eQTL.
Conflict of interest statement
Ethics approval and consent to participate
All studies were carried out according to institutional guidelines, and with appropriate informed consent from participants or next of kin (in case of donor heart samples). Institutional ethics committees of the clinical centers where the cardiac samples were collected reviewed and approved all protocols. The ethical review boards of University of Szeged (Ethical Review Board of the University of Szeged Medical Center; Szeged, Hungary), Vanderbilt University (Institutional Review Board of Vanderbilt University School of Medicine; Nashville, USA, IRB#100664), University of Miami (Institutional Review Board of the University of Miami School of Medicine; Miami, USA, protocol # 20010028), and the University of Sydney (Human Research Ethics Committee (HREC), Project Title: The Sydney Human Heart Tissue Bank (SHB), project number 2012/2814; Sydney, Australia) approved procurement and handling of the human donor cardiac material. DCM tissue studies complied with UK Human Tissue Act guidelines and were carried out with approval from the Royal Brompton and Harefield local ethical review committee and the National Research Ethics Service Committee South Central, Hampshire B (reference 09/H0504/104). Investigations conformed to the principles outlined in the Helsinki Declaration of the World Medical Association. All data were analyzed anonymously.
Competing interests
The authors declare that they have no competing interests.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

