Molecular Tools for the Detection and Deduction of Azole Antifungal Drug Resistance Phenotypes in Aspergillus Species
- PMID: 28903985
- PMCID: PMC5608879
- DOI: 10.1128/CMR.00095-16
Molecular Tools for the Detection and Deduction of Azole Antifungal Drug Resistance Phenotypes in Aspergillus Species
Abstract
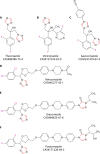
The incidence of azole resistance in Aspergillus species has increased over the past years, most importantly for Aspergillus fumigatus. This is partially attributable to the global spread of only a few resistance alleles through the environment. Secondary resistance is a significant clinical concern, as invasive aspergillosis with drug-susceptible strains is already difficult to treat, and exclusion of azole-based antifungals from prophylaxis or first-line treatment of invasive aspergillosis in high-risk patients would dramatically limit drug choices, thus increasing mortality rates for immunocompromised patients. Management options for invasive aspergillosis caused by azole-resistant A. fumigatus strains were recently reevaluated by an international expert panel, which concluded that drug resistance testing of cultured isolates is highly indicated when antifungal therapy is intended. In geographical regions with a high environmental prevalence of azole-resistant strains, initial therapy should be guided by such analyses. More environmental and clinical screening studies are therefore needed to generate the local epidemiologic data if such measures are to be implemented on a sound basis. Here we propose a first workflow for evaluating isolates from screening studies, and we compile the MIC values correlating with individual amino acid substitutions in the products of cyp51 genes for interpretation of DNA sequencing data, especially in the absence of cultured isolates.
Keywords: Aspergillus flavus; Aspergillus fumigatus; Aspergillus niger; Aspergillus terreus; azole drug resistance; cyp51A; cyp51B; cyp51C; diagnostics; efflux; tandem repeats.
Copyright © 2017 American Society for Microbiology.
Figures












References
-
- van de Peppel RJ, von dem Borne PA, le Cessie S, de Boer MGJ. 2017. A new time-dependent approach for assessment of the impact of invasive aspergillosis shows effect on short- but not on long-term survival of patients with AML or high-risk MDS. Bone Marrow Transplant 52:883–888. doi: 10.1038/bmt.2017.71. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

