Identification of sites of 2'-O-methylation vulnerability in human ribosomal RNAs by systematic mapping
- PMID: 28904332
- PMCID: PMC5597630
- DOI: 10.1038/s41598-017-09734-9
Identification of sites of 2'-O-methylation vulnerability in human ribosomal RNAs by systematic mapping
Abstract
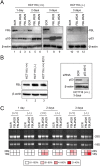
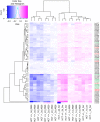
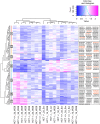
Ribosomal RNA modifications are important in optimizing ribosome function. Sugar 2'-O-methylation performed by fibrillarin-associated box C/D antisense guide snoRNAs impacts all steps of translation, playing a role in disease etiology (cancer). As it renders adjacent phosphodiester bonds resistant to alkaline treatment, 2'-O-methylation can be monitored qualitatively and quantitatively by applying next-generation sequencing to fragments of randomly cleaved RNA. We remapped all sites of 2'-O-methylation in human rRNAs in two isogenic diploid cell lines, one producing and one not producing the antitumor protein p53. We identified sites naturally modified only partially (confirming the existence in cells of compositionally distinct ribosomes with potentially specialized functions) and sites whose 2'-O-methylation is sensitive to p53. We mapped sites particularly vulnerable to a reduced level of the methyltransferase fibrillarin. The remarkable fact that these are largely sites of natural hypomodification provides initial insights into the mechanism of partial RNA modification. Sites where methylation appeared vulnerable lie peripherally on the 3-D structure of the ribosomal subunits, whereas the numerous modifications present at the core of the subunits, where the functional centers lie, appeared robustly made. We suggest that vulnerable sites of 2'-O-methylation are highly likely to undergo specific regulation during normal and pathological processes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

