Toxoplasma Modulates Signature Pathways of Human Epilepsy, Neurodegeneration & Cancer
- PMID: 28904337
- PMCID: PMC5597608
- DOI: 10.1038/s41598-017-10675-6
Toxoplasma Modulates Signature Pathways of Human Epilepsy, Neurodegeneration & Cancer
Erratum in
-
Publisher Correction: Toxoplasma Modulates Signature Pathways of Human Epilepsy, Neurodegeneration & Cancer.Sci Rep. 2019 May 28;9(1):8110. doi: 10.1038/s41598-019-44545-0. Sci Rep. 2019. PMID: 31133744 Free PMC article.
Abstract
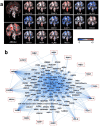
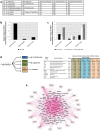
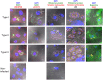
One third of humans are infected lifelong with the brain-dwelling, protozoan parasite, Toxoplasma gondii. Approximately fifteen million of these have congenital toxoplasmosis. Although neurobehavioral disease is associated with seropositivity, causality is unproven. To better understand what this parasite does to human brains, we performed a comprehensive systems analysis of the infected brain: We identified susceptibility genes for congenital toxoplasmosis in our cohort of infected humans and found these genes are expressed in human brain. Transcriptomic and quantitative proteomic analyses of infected human, primary, neuronal stem and monocytic cells revealed effects on neurodevelopment and plasticity in neural, immune, and endocrine networks. These findings were supported by identification of protein and miRNA biomarkers in sera of ill children reflecting brain damage and T. gondii infection. These data were deconvoluted using three systems biology approaches: "Orbital-deconvolution" elucidated upstream, regulatory pathways interconnecting human susceptibility genes, biomarkers, proteomes, and transcriptomes. "Cluster-deconvolution" revealed visual protein-protein interaction clusters involved in processes affecting brain functions and circuitry, including lipid metabolism, leukocyte migration and olfaction. Finally, "disease-deconvolution" identified associations between the parasite-brain interactions and epilepsy, movement disorders, Alzheimer's disease, and cancer. This "reconstruction-deconvolution" logic provides templates of progenitor cells' potentiating effects, and components affecting human brain parasitism and diseases.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

