Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection
- PMID: 28904745
- PMCID: PMC5591563
- DOI: 10.1186/s13578-017-0174-2
Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection
Abstract
Background: The main approach to treat HIV-1 infection is combination antiretroviral therapy (cART). Although cART is effective in reducing HIV-1 viral load and controlling disease progression, it has many side effects, and is expensive for HIV-1 infected patients who must remain on lifetime treatment. HIV-1 gene therapy has drawn much attention as studies of genome editing tools have progressed. For example, zinc finger nucleases (ZFN), transcription activator like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 have been utilized to successfully disrupt the HIV-1 co-receptors CCR5 or CXCR4, thereby restricting HIV-1 infection. However, the effects of simultaneous genome editing of CXCR4 and CCR5 by CRISPR-Cas9 in blocking HIV-1 infection in primary CD4+ T cells has been rarely reported. Furthermore, combination of different target sites of CXCR4 and CCR5 for disruption also need investigation.
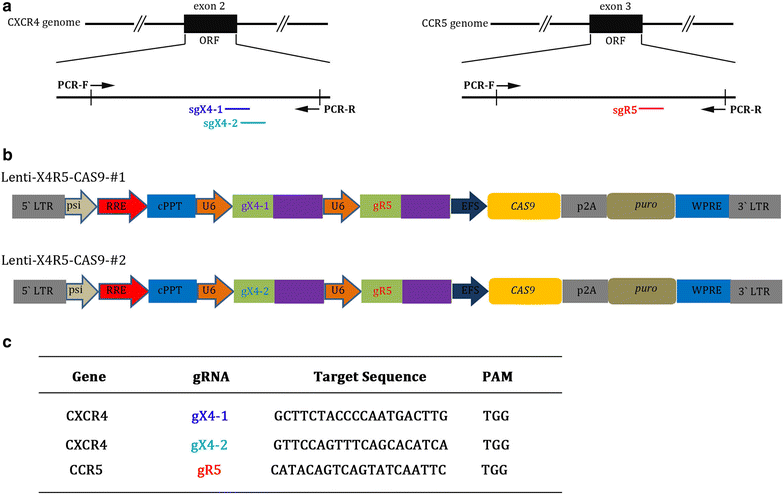
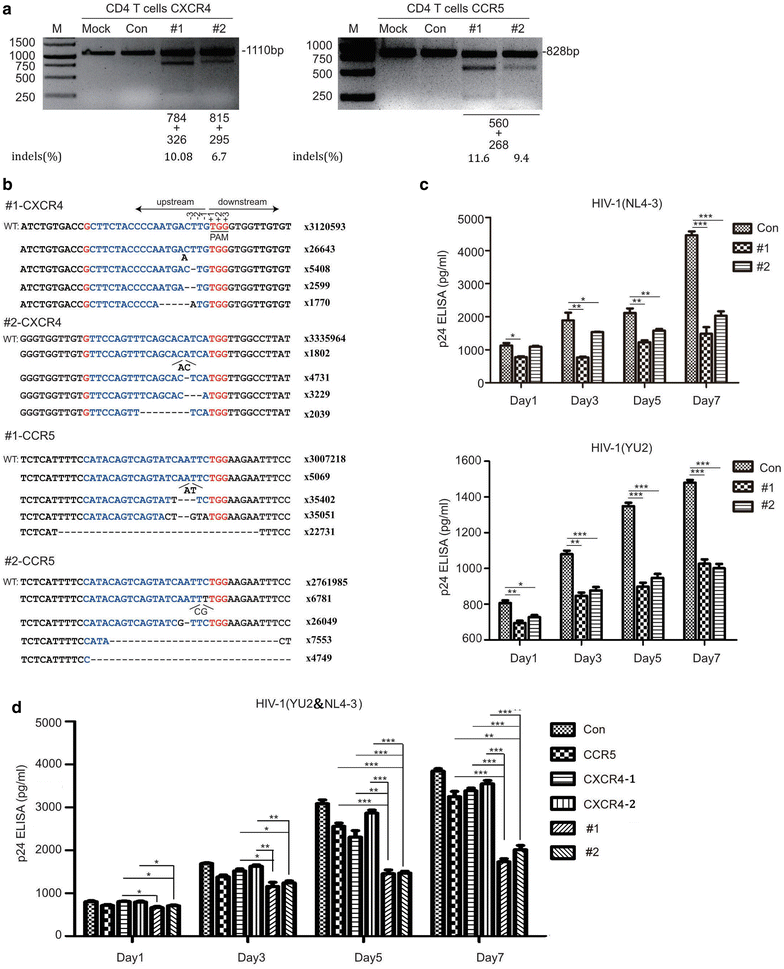
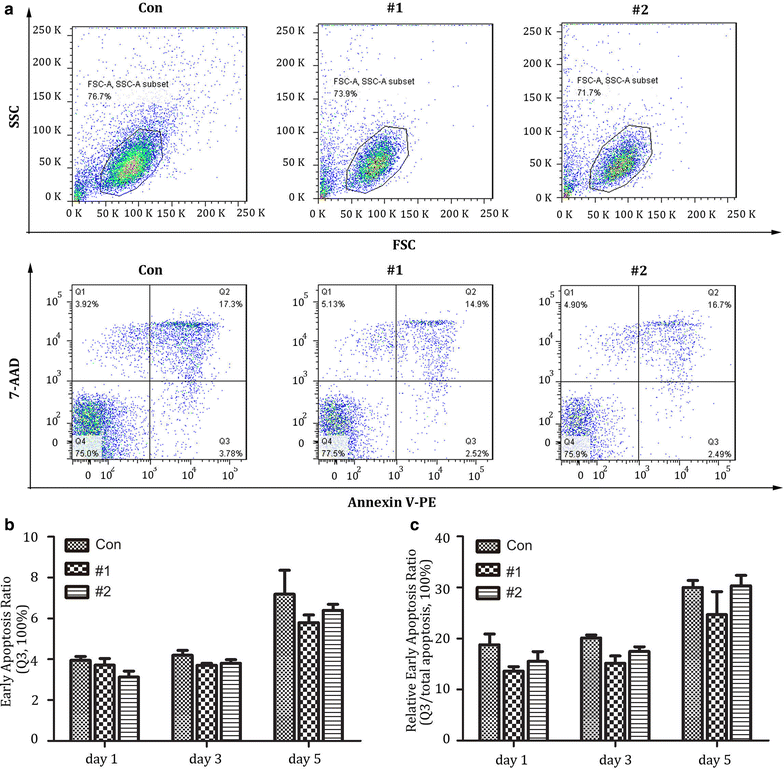
Results: In this report, we designed two different gRNA combinations targeting both CXCR4 and CCR5, in a single vector. The CRISPR-sgRNAs-Cas9 could successfully induce editing of CXCR4 and CCR5 genes in various cell lines and primary CD4+ T cells. Using HIV-1 challenge assays, we demonstrated that CXCR4-tropic or CCR5-tropic HIV-1 infections were significantly reduced in CXCR4- and CCR5-modified cells, and the modified cells exhibited a selective advantage over unmodified cells during HIV-1 infection. The off-target analysis showed that no non-specific editing was identified in all predicted sites. In addition, apoptosis assays indicated that simultaneous disruption of CXCR4 and CCR5 in primary CD4+ T cells by CRISPR-Cas9 had no obvious cytotoxic effects on cell viability.
Conclusions: Our results suggest that simultaneous genome editing of CXCR4 and CCR5 by CRISPR-Cas9 can potentially provide an effective and safe strategy towards a functional cure for HIV-1 infection.
Keywords: AIDS; CCR5 and CXCR4 simultaneous; CRISPR-Cas9; HIV-1.
Figures




References
-
- Update on acquired immune deficiency syndrome (AIDS)–United States. MMWR. 1982;31(37):507-8, 13-4. (Epub 1982/09/24). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

