Identification and Characterization of Domesticated Bacterial Transposases
- PMID: 28910967
- PMCID: PMC5581495
- DOI: 10.1093/gbe/evx146
Identification and Characterization of Domesticated Bacterial Transposases
Abstract
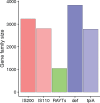
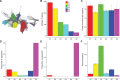
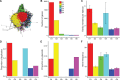
Selfish genetic elements, such as insertion sequences and transposons are found in most genomes. Transposons are usually identifiable by their high copy number within genomes. In contrast, REP-associated tyrosine transposases (RAYTs), a recently described class of bacterial transposase, are typically present at just one copy per genome. This suggests that RAYTs no longer copy themselves and thus they no longer function as a typical transposase. Motivated by this possibility we interrogated thousands of fully sequenced bacterial genomes in order to determine patterns of RAYT diversity, their distribution across chromosomes and accessory elements, and rate of duplication. RAYTs encompass exceptional diversity and are divisible into at least five distinct groups. They possess features more similar to housekeeping genes than insertion sequences, are predominantly vertically transmitted and have persisted through evolutionary time to the point where they are now found in 24% of all species for which at least one fully sequenced genome is available. Overall, the genomic distribution of RAYTs suggests that they have been coopted by host genomes to perform a function that benefits the host cell.
Keywords: IS200; RAYTs; REPINs; domesticated transposons; selfish genetic elements.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Single-strand DNA processing: phylogenomics and sequence diversity of a superfamily of potential prokaryotic HuH endonucleases.BMC Genomics. 2018 Jun 19;19(1):475. doi: 10.1186/s12864-018-4836-1. BMC Genomics. 2018. PMID: 29914351 Free PMC article.
-
Identification and characterization of repetitive extragenic palindromes (REP)-associated tyrosine transposases: implications for REP evolution and dynamics in bacterial genomes.BMC Genomics. 2010 Jan 19;11:44. doi: 10.1186/1471-2164-11-44. BMC Genomics. 2010. PMID: 20085626 Free PMC article.
-
Evolution of REP diversity: a comparative study.BMC Genomics. 2013 Jun 10;14:385. doi: 10.1186/1471-2164-14-385. BMC Genomics. 2013. PMID: 23758774 Free PMC article.
-
Miniature inverted-repeat transposable elements: discovery, distribution, and activity.Genome. 2013 Sep;56(9):475-86. doi: 10.1139/gen-2012-0174. Epub 2013 Mar 8. Genome. 2013. PMID: 24168668 Review.
-
Bacterial repetitive extragenic palindromic sequences are DNA targets for Insertion Sequence elements.BMC Genomics. 2006 Mar 24;7:62. doi: 10.1186/1471-2164-7-62. BMC Genomics. 2006. PMID: 16563168 Free PMC article. Review.
Cited by
-
Discovering Complete Quasispecies in Bacterial Genomes.Genetics. 2017 Aug;206(4):2149-2157. doi: 10.1534/genetics.117.201160. Epub 2017 Jun 19. Genetics. 2017. PMID: 28630115 Free PMC article.
-
The ISPpu9 insertion sequence of Pseudomonas putida KT2440 generates various circular intermediates enabling modular transposition.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf668. doi: 10.1093/nar/gkaf668. Nucleic Acids Res. 2025. PMID: 40705920 Free PMC article.
-
How sequence populations persist inside bacterial genomes.Genetics. 2021 Apr 15;217(4):iyab027. doi: 10.1093/genetics/iyab027. Genetics. 2021. PMID: 33724360 Free PMC article.
-
Single-strand DNA processing: phylogenomics and sequence diversity of a superfamily of potential prokaryotic HuH endonucleases.BMC Genomics. 2018 Jun 19;19(1):475. doi: 10.1186/s12864-018-4836-1. BMC Genomics. 2018. PMID: 29914351 Free PMC article.
-
Expression of the ISPpu9 transposase of Pseudomonas putida KT2440 is regulated by two small RNAs and the secondary structure of the mRNA 5'-untranslated region.Nucleic Acids Res. 2021 Sep 20;49(16):9211-9228. doi: 10.1093/nar/gkab672. Nucleic Acids Res. 2021. PMID: 34379788 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

