DNA-binding properties of FOXP3 transcription factor
- PMID: 28910978
- PMCID: PMC6276865
- DOI: 10.1093/abbs/gmx079
DNA-binding properties of FOXP3 transcription factor
Abstract
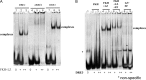
FOXP3, a lineage-specific forkhead (FKH) transcription factor, plays essential roles in the development and function of regulatory T cells. However, the DNA-binding properties of FOXP3 are not well understood. In this study, FOXP3 fragments containing different domains were purified, and their DNA-binding properties were investigated using electrophoretic mobility shift assay and isothermal titration calorimetry (ITC). Both the FKH and leucine-zipper domains were required for optimal DNA binding for FOXP3. FOXP3 protein not only binds with DNA sequences containing one FKH consensus sequence, but also binds with DNA sequences with two direct repeats of consensus sequences separated by three-nucleotides (DRE3). Our results shed lights on the mechanisms by which FOXP3 recognizes cognate DNA elements, and would facilitate further structural and functional studies of FOXP3.
Keywords: DNA binding; FOXP3; electrophoretic mobility shift assay; isothermal titration calorimetry; protein purification.
© The Author 2017. Published by Oxford University Press on behalf of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures





Similar articles
-
Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3.PLoS One. 2009 Dec 1;4(12):e8109. doi: 10.1371/journal.pone.0008109. PLoS One. 2009. PMID: 19956618 Free PMC article.
-
A Key Evolutionary Mutation Enhances DNA Binding of the FOXP2 Forkhead Domain.Biochemistry. 2016 Apr 5;55(13):1959-67. doi: 10.1021/acs.biochem.5b01271. Epub 2016 Mar 18. Biochemistry. 2016. PMID: 26950495
-
The crystal structure of human forkhead box N1 in complex with DNA reveals the structural basis for forkhead box family specificity.J Biol Chem. 2020 Mar 6;295(10):2948-2958. doi: 10.1074/jbc.RA119.010365. Epub 2019 Dec 30. J Biol Chem. 2020. PMID: 31914405 Free PMC article.
-
Structural basis for DNA recognition by FOXO proteins.Biochim Biophys Acta. 2011 Nov;1813(11):1946-53. doi: 10.1016/j.bbamcr.2010.11.025. Epub 2010 Dec 10. Biochim Biophys Acta. 2011. PMID: 21146564 Review.
-
Experimental approaches to investigate biophysical interactions between homeodomain transcription factors and DNA.Biochim Biophys Acta Gene Regul Mech. 2025 Mar;1868(1):195074. doi: 10.1016/j.bbagrm.2024.195074. Epub 2024 Dec 5. Biochim Biophys Acta Gene Regul Mech. 2025. PMID: 39644990 Free PMC article. Review.
Cited by
-
Identification of a group of 9-amino-acridines that selectively downregulate regulatory T cell functions through FoxP3.iScience. 2025 Jan 31;28(3):111931. doi: 10.1016/j.isci.2025.111931. eCollection 2025 Mar 21. iScience. 2025. PMID: 40034859 Free PMC article.
-
Structural basis for DNA recognition by FOXC2.Nucleic Acids Res. 2019 Apr 23;47(7):3752-3764. doi: 10.1093/nar/gkz077. Nucleic Acids Res. 2019. PMID: 30722065 Free PMC article.
-
Post-Translational Regulations of Foxp3 in Treg Cells and Their Therapeutic Applications.Front Immunol. 2021 Apr 12;12:626172. doi: 10.3389/fimmu.2021.626172. eCollection 2021. Front Immunol. 2021. PMID: 33912156 Free PMC article. Review.
-
The Importance of the Transcription Factor Foxp3 in the Development of Primary Immunodeficiencies.J Clin Med. 2022 Feb 11;11(4):947. doi: 10.3390/jcm11040947. J Clin Med. 2022. PMID: 35207219 Free PMC article. Review.
-
IGF1R activates FOXP3-β-catenin signaling to promote breast cancer development.Breast Cancer Res Treat. 2025 Jun;211(2):467-478. doi: 10.1007/s10549-025-07663-0. Epub 2025 Mar 7. Breast Cancer Res Treat. 2025. PMID: 40055251
References
-
- Zheng Y, Rudensky A. Foxp3 in control of the regulatory T cell lineage. Nat Immunol 2007, 8: 457–462. - PubMed
-
- Park J, Ko JS, Shin Y, Cho JY, Oh H, Bothwell AM, Lee S. Intranuclear interactomic inhibition of FoxP3 suppresses functions of Treg cells. Biochem Biophys Res Commun 2014, 451: 1–7. - PubMed
-
- Joly A, Liu S, Dahlberg CIM, Mailer RKW, Westerberg LS, Andersson J. Foxp3 lacking exons 2 and 7 is unable to confer suppressive ability to regulatory T cells in vivo. J Autoimmun 2015, 63: 23–30. - PubMed
-
- Buckner JH, Ziegler SF. Functional analysis of FOXP3. Ann N Y Acad Sci 2008, 1143: 151–169. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

