Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
- PMID: 28911094
- PMCID: PMC5587774
- DOI: 10.1093/nar/gkx547
Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
Abstract
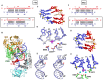
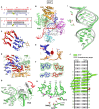
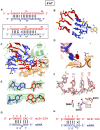
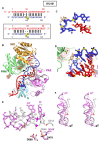
We have undertaken a systematic structural study of Thermus thermophilus Argonaute (TtAgo) ternary complexes containing single-base bulges positioned either within the seed segment of the guide or target strands and at the cleavage site. Our studies establish that single-base bulges 7T8, 5A6 and 4A5 on the guide strand are stacked-into the duplex, with conformational changes localized to the bulge site, thereby having minimal impact on the cleavage site. By contrast, single-base bulges 6'U7' and 6'A7' on the target strand are looped-out of the duplex, with the resulting conformational transitions shifting the cleavable phosphate by one step. We observe a stable alignment for the looped-out 6'N7' bulge base, which stacks on the unpaired first base of the guide strand, with the looped-out alignment facilitated by weakened Watson-Crick and reversed non-canonical flanking pairs. These structural studies are complemented by cleavage assays that independently monitor the impact of bulges on TtAgo-mediated cleavage reaction.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Dykxhoorn D.M., Novina C.D., Sharp P.A.. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003; 4:457–467. - PubMed
-
- Baulcombe D. RNA silencing in plants. Nature. 2004; 431:356–363. - PubMed
-
- Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005; 122:17–20. - PubMed
-
- Rana T.M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:23–36. - PubMed
-
- Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

