The SmAP2 RNA binding motif in the 3'UTR affects mRNA stability in the crenarchaeum Sulfolobus solfataricus
- PMID: 28911098
- PMCID: PMC5587771
- DOI: 10.1093/nar/gkx581
The SmAP2 RNA binding motif in the 3'UTR affects mRNA stability in the crenarchaeum Sulfolobus solfataricus
Abstract
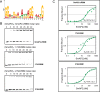
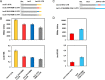
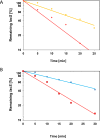
Sm and Sm-like proteins represent an evolutionarily conserved family with key roles in RNA metabolism in Pro- and Eukaryotes. In this study, a collection of 53 mRNAs that co-purified with Sulfolobus solfataricus (Sso) SmAP2 were surveyed for a specific RNA binding motif (RBM). SmAP2 was shown to bind with high affinity to the deduced consensus RNA binding motif (SmAP2-cRBM) in vitro. Residues in SmAP2 interacting with the SmAP2-cRBM were mapped by UV-induced crosslinking in combination with mass-spectrometry, and verified by mutational analyses. The RNA-binding site on SmAP2 includes a modified uracil binding pocket containing a unique threonine (T40) located on the L3 face and a second residue, K25, located in the pore. To study the function of the SmAP2-RBM in vivo, three authentic RBMs were inserted in the 3'UTR of a lacS reporter gene. The presence of the SmAP2-RBM in the reporter-constructs resulted in decreased LacS activity and reduced steady state levels of lacS mRNA. Moreover, the presence of the SmAP2-cRBM in and the replacement of the lacS 3'UTR with that of Sso2194 encompassing a SmAP2-RBM apparently impacted on the stability of the chimeric transcripts. These results are discussed in light of the function(s) of eukaryotic Lsm proteins in RNA turnover.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.J Biomol Struct Dyn. 2017 Jun;35(8):1615-1628. doi: 10.1080/07391102.2016.1189849. Epub 2016 Aug 1. J Biomol Struct Dyn. 2017. PMID: 27187760
-
The SmAP1/2 proteins of the crenarchaeon Sulfolobus solfataricus interact with the exosome and stimulate A-rich tailing of transcripts.Nucleic Acids Res. 2017 Jul 27;45(13):7938-7949. doi: 10.1093/nar/gkx437. Nucleic Acids Res. 2017. PMID: 28520934 Free PMC article.
-
Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus.EMBO Rep. 2013 Jun;14(6):527-33. doi: 10.1038/embor.2013.47. Epub 2013 Apr 12. EMBO Rep. 2013. PMID: 23579342 Free PMC article.
-
Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function.RNA Biol. 2013 Apr;10(4):636-51. doi: 10.4161/rna.24538. Epub 2013 Apr 1. RNA Biol. 2013. PMID: 23579284 Free PMC article. Review.
-
Structure and function of the archaeal exosome.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):623-35. doi: 10.1002/wrna.1234. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24789718 Review.
Cited by
-
AU-Rich Long 3' Untranslated Region Regulates Gene Expression in Bacteria.Front Microbiol. 2018 Dec 12;9:3080. doi: 10.3389/fmicb.2018.03080. eCollection 2018. Front Microbiol. 2018. PMID: 30619162 Free PMC article.
-
Comparative CRISPR type III-based knockdown of essential genes in hyperthermophilic Sulfolobales and the evasion of lethal gene silencing.RNA Biol. 2021 Mar;18(3):421-434. doi: 10.1080/15476286.2020.1813411. Epub 2020 Sep 21. RNA Biol. 2021. PMID: 32957821 Free PMC article.
-
Novel roles for Sm-class RNAs in the regulation of gene expression.RNA Biol. 2018;15(7):856-862. doi: 10.1080/15476286.2018.1467176. Epub 2018 Jul 9. RNA Biol. 2018. PMID: 29895222 Free PMC article. Review.
-
A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius.Nucleic Acids Res. 2018 May 18;46(9):4794-4806. doi: 10.1093/nar/gky144. Nucleic Acids Res. 2018. PMID: 29529252 Free PMC article.
References
-
- Collins B.M., Harrop S.J., Kornfeld G.D., Dawes I.W., Curmi P.M., Mabbutt B.C.. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001; 309:915–923. - PubMed
-
- Törö I., Basquin J., Teo-Dreher H., Suck D.. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J. Mol. Biol. 2002; 320:129–142. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

