Enhancement of mouse hematopoietic stem/progenitor cell function via transient gene delivery using integration-deficient lentiviral vectors
- PMID: 28911908
- PMCID: PMC5731634
- DOI: 10.1016/j.exphem.2017.09.003
Enhancement of mouse hematopoietic stem/progenitor cell function via transient gene delivery using integration-deficient lentiviral vectors
Abstract
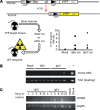
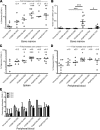
Integration-deficient lentiviruses (IdLVs) deliver genes effectively to tissues but are lost rapidly from dividing cells. This property can be harnessed to express transgenes transiently to manipulate cell biology. Here, we demonstrate the utility of short-term gene expression to improve functional potency of hematopoietic stem and progenitor cells (HSPCs) during transplantation by delivering HOXB4 and Angptl3 using IdLVs to enhance the engraftment of HSPCs. Constitutive overexpression of either of these genes is likely to be undesirable, but the transient nature of IdLVs reduces this risk and those associated with unsolicited gene expression in daughter cells. Transient expression led to increased multilineage hematopoietic engraftment in in vivo competitive repopulation assays without the side effects reported in constitutive overexpression models. Adult stem cell fate has not been programmed previously using IdLVs, but we demonstrate that these transient gene expression tools can produce clinically relevant alterations or be applied to investigate basic biology.
Copyright © 2018 ISEH – Society for Hematology and Stem Cells. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Specific transgene expression in human and mouse CD4+ cells using lentiviral vectors with regulatory sequences from the CD4 gene.Blood. 2003 May 1;101(9):3416-23. doi: 10.1182/blood-2002-02-0578. Epub 2003 Jan 2. Blood. 2003. PMID: 12511423
-
Increased Engraftment of Human Short Term Repopulating Hematopoietic Cells in NOD/SCID/IL2rγnull Mice by Lentiviral Expression of NUP98-HOXA10HD.PLoS One. 2016 Jan 13;11(1):e0147059. doi: 10.1371/journal.pone.0147059. eCollection 2016. PLoS One. 2016. PMID: 26761813 Free PMC article.
-
Forced expression of HoxB4 enhances hematopoietic differentiation by human embryonic stem cells.Mol Cells. 2008 Jun 30;25(4):487-93. Epub 2008 May 29. Mol Cells. 2008. PMID: 18511880
-
Advances in lentiviral vector design for gene-modification of hematopoietic stem cells.Curr Opin Biotechnol. 2002 Oct;13(5):429-36. doi: 10.1016/s0958-1669(02)00346-4. Curr Opin Biotechnol. 2002. PMID: 12459333 Review.
-
[Ex vivo expansion of human hematopoietic stem cells by passive transduction of the HOXB4 homeoprotein].J Soc Biol. 2006;200(3):235-41. doi: 10.1051/jbio:2006027. J Soc Biol. 2006. PMID: 17417138 Review. French.
Cited by
-
Designing an Engineered Construct Gene Sensitive to Carbohydrate In-vitro and Candidate for Human Insulin Gene Therapy In-vivo.Iran J Pharm Res. 2019 Fall;18(4):2111-2116. doi: 10.22037/ijpr.2019.14650.12567. Iran J Pharm Res. 2019. PMID: 32184874 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- MR/R015325/1/MRC_/Medical Research Council/United Kingdom
- MR/P026494/1/MRC_/Medical Research Council/United Kingdom
- R01 DK062757/DK/NIDDK NIH HHS/United States
- BB/I00212X/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G1000709/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

