Assessing the mechanisms of cholesteryl ester transfer protein inhibitors
- PMID: 28911944
- PMCID: PMC6239860
- DOI: 10.1016/j.bbalip.2017.09.004
Assessing the mechanisms of cholesteryl ester transfer protein inhibitors
Abstract
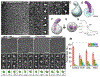
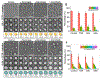
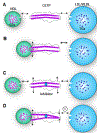
Cholesteryl ester transfer protein (CETP) inhibitors are a new class of therapeutics for dyslipidemia that simultaneously improve two major cardiovascular disease (CVD) risk factors: elevated low-density lipoprotein (LDL) cholesterol and decreased high-density lipoprotein (HDL) cholesterol. However, the detailed molecular mechanisms underlying their efficacy are poorly understood, as are any potential mechanistic differences among the drugs in this class. Herein, we used electron microscopy (EM) to investigate the effects of three of these agents (Torcetrapib, Dalcetrapib and Anacetrapib) on CETP structure, CETP-lipoprotein complex formation and CETP-mediated cholesteryl ester (CE) transfer. We found that although none of these inhibitors altered the structure of CETP or the conformation of CETP-lipoprotein binary complexes, all inhibitors, especially Torcetrapib and Anacetrapib, increased the binding ratios of the binary complexes (e.g., HDL-CETP and LDL-CETP) and decreased the binding ratios of the HDL-CETP-LDL ternary complexes. The findings of more binary complexes and fewer ternary complexes reflect a new mechanism of inhibition: one distal end of CETP bound to the first lipoprotein would trigger a conformational change at the other distal end, thus resulting in a decreased binding ratio to the second lipoprotein and a degraded CE transfer rate among lipoproteins. Thus, we suggest a new inhibitor design that should decrease the formation of both binary and ternary complexes. Decreased concentrations of the binary complex may prevent the inhibitor was induced into cell by the tight binding of binary complexes during lipoprotein metabolism in the treatment of CVD.
Keywords: Anacetrapib; CETP; CETP bound to HDL; CETP inhibitor; Cholesteryl ester transfer protein; Dalcetrapib; Electron microscopy; HDL; LDL; Torcetrapib; VLDL.
Published by Elsevier B.V.
Figures



Similar articles
-
Biochemical characterization of cholesteryl ester transfer protein inhibitors.J Lipid Res. 2010 Sep;51(9):2739-52. doi: 10.1194/jlr.M007468. Epub 2010 May 10. J Lipid Res. 2010. PMID: 20458119 Free PMC article.
-
Cholesteryl ester transfer-protein modulator and inhibitors and their potential for the treatment of cardiovascular diseases.Vasc Health Risk Manag. 2012;8:323-31. doi: 10.2147/VHRM.S25238. Epub 2012 May 15. Vasc Health Risk Manag. 2012. PMID: 22661899 Free PMC article. Review.
-
Future of cholesteryl ester transfer protein (CETP) inhibitors: a pharmacological perspective.Clin Pharmacokinet. 2013 Aug;52(8):615-26. doi: 10.1007/s40262-013-0071-8. Clin Pharmacokinet. 2013. PMID: 23658137 Free PMC article.
-
Cholesteryl ester transfer protein: An enigmatic pharmacology - Antagonists and agonists.Atherosclerosis. 2018 Nov;278:286-298. doi: 10.1016/j.atherosclerosis.2018.09.035. Epub 2018 Oct 1. Atherosclerosis. 2018. PMID: 30347344 Review.
-
On- and off-target pharmacology of torcetrapib: current understanding and implications for the structure activity relationships (SAR), discovery and development of cholesteryl ester-transfer protein (CETP) inhibitors.Drugs. 2012 Mar 5;72(4):491-507. doi: 10.2165/11599310-000000000-00000. Drugs. 2012. PMID: 22356288 Review.
Cited by
-
Optimized Negative-Staining Protocol for Lipid-Protein Interactions Investigated by Electron Microscopy.Methods Mol Biol. 2019;2003:163-173. doi: 10.1007/978-1-4939-9512-7_8. Methods Mol Biol. 2019. PMID: 31218618 Free PMC article.
-
In Silico Strategies for Characterizing Inner Cavities of Lipid-Binding Proteins.Methods Mol Biol. 2025;2888:305-320. doi: 10.1007/978-1-0716-4318-1_20. Methods Mol Biol. 2025. PMID: 39699739
-
Structural basis of the lipid transfer mechanism of phospholipid transfer protein (PLTP).Biochim Biophys Acta Mol Cell Biol Lipids. 2018 Sep;1863(9):1082-1094. doi: 10.1016/j.bbalip.2018.06.001. Epub 2018 Jun 5. Biochim Biophys Acta Mol Cell Biol Lipids. 2018. PMID: 29883800 Free PMC article.
-
Single-Molecule 3D Images of "Hole-Hole" IgG1 Homodimers by Individual-Particle Electron Tomography.Sci Rep. 2019 Jun 20;9(1):8864. doi: 10.1038/s41598-019-44978-7. Sci Rep. 2019. PMID: 31221961 Free PMC article.
-
Chemical Synthesis, Biological Evaluation, and Cheminformatics Analysis of a Group of Chlorinated Diaryl Sulfonamides: Promising Inhibitors of Cholesteryl Ester Transfer Protein.Curr Comput Aided Drug Des. 2025;21(5):694-707. doi: 10.2174/0115734099292078240218095540. Curr Comput Aided Drug Des. 2025. PMID: 38424428
References
-
- Barter PJ, Brewer HB Jr., Chapman MJ, Hennekens CH, Rader DJ, Tall AR, Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis, Arterioscler. Thromb. Vasc. Biol 23 (2003) 160–167. - PubMed
-
- Inazu A, Brown ML, Hesler CB, Agellon LB, Koizumi J, Takata K, Maruhama Y, Mabuchi H, Tall AR, Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation, N. Engl. J. Med 323 (1990) 1234–1238. - PubMed
-
- Brown ML, Inazu A, Hesler CB, Agellon LB, Mann C, Whitlock ME, Marcel YL, Milne RW, Koizumi J, Mabuchi H, et al., Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins, Nature 342 (1989) 448–451. - PubMed
-
- Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B, Investigators I, Effects of torcetrapib in patients at high risk for coronary events, N. Engl. J. Med 357 (2007) 2109–2122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

