Systematic Mutagenesis of Serine Hydroxymethyltransferase Reveals an Essential Role in Nematode Resistance
- PMID: 28912378
- PMCID: PMC5664460
- DOI: 10.1104/pp.17.00553
Systematic Mutagenesis of Serine Hydroxymethyltransferase Reveals an Essential Role in Nematode Resistance
Abstract
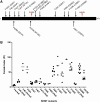
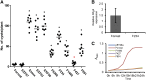
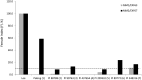
Rhg4 is a major genetic locus that contributes to soybean cyst nematode (SCN) resistance in the Peking-type resistance of soybean (Glycine max), which also requires the rhg1 gene. By map-based cloning and functional genomic approaches, we previously showed that the Rhg4 gene encodes a predicted cytosolic serine hydroxymethyltransferase (GmSHMT08); however, the novel gain of function of GmSHMT08 in SCN resistance remains to be characterized. Using a forward genetic screen, we identified an allelic series of GmSHMT08 mutants that shed new light on the mechanistic aspects of GmSHMT08-mediated resistance. The new mutants provide compelling genetic evidence that Peking-type rhg1 resistance in cv Forrest is fully dependent on the GmSHMT08 gene and demonstrates that this resistance is mechanistically different from the PI 88788-type of resistance that only requires rhg1 We also demonstrated that rhg1-a from cv Forrest, although required, does not exert selection pressure on the nematode to shift from HG type 7, which further validates the bigenic nature of this resistance. Mapping of the identified mutations onto the SHMT structural model uncovered key residues for structural stability, ligand binding, enzyme activity, and protein interactions, suggesting that GmSHMT08 has additional functions aside from its main enzymatic role in SCN resistance. Lastly, we demonstrate the functionality of the GmSHMT08 SCN resistance gene in a transgenic soybean plant.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Appaji Rao N, Ambili M, Jala VR, Subramanya HS, Savithri HS (2003) Structure-function relationship in serine hydroxymethyltransferase. Biochim Biophys Acta 1647: 24–29 - PubMed
-
- Blakely RL. (1969) The biochemistry of folic acid and related pteridines. Frontiers in Biology 13: 189–218
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

