Adaptation of Surface-Associated Bacteria to the Open Ocean: A Genomically Distinct Subpopulation of Phaeobacter gallaeciensis Colonizes Pacific Mesozooplankton
- PMID: 28912769
- PMCID: PMC5583230
- DOI: 10.3389/fmicb.2017.01659
Adaptation of Surface-Associated Bacteria to the Open Ocean: A Genomically Distinct Subpopulation of Phaeobacter gallaeciensis Colonizes Pacific Mesozooplankton
Abstract
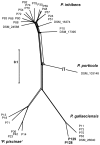
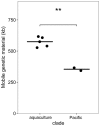
The marine Roseobacter group encompasses numerous species which occupy a large variety of ecological niches. However, members of the genus Phaeobacter are specifically adapted to a surface-associated lifestyle and have so far been found nearly exclusively in disjunct, man-made environments including shellfish and fish aquacultures, as well as harbors. Therefore, the possible natural habitats, dispersal and evolution of Phaeobacter spp. have largely remained obscure. Applying a high-throughput cultivation strategy along a longitudinal Pacific transect, the present study revealed for the first time a widespread natural occurrence of Phaeobacter in the marine pelagial. These bacteria were found to be specifically associated to mesoplankton where they constitute a small but detectable proportion of the bacterial community. The 16S rRNA gene sequences of 18 isolated strains were identical to that of Phaeobacter gallaeciensis DSM26640T but sequences of internal transcribed spacer and selected genomes revealed that the strains form a distinct clade within P. gallaeciensis. The genomes of the Pacific and the aquaculture strains were highly conserved and had a fraction of the core genome of 89.6%, 80 synteny breakpoints, and differed 2.2% in their nucleotide sequences. Diversification likely occurred through neutral mutations. However, the Pacific strains exclusively contained two active Type I restriction modification systems which is commensurate with a reduced acquisition of mobile elements in the Pacific clade. The Pacific clade of P. gallaeciensis also acquired a second, homolog phosphonate transport system compared to all other P. gallaeciensis. Our data indicate that a previously unknown, distinct clade of P. gallaeciensis acquired a limited number of clade-specific genes that were relevant for its association with mesozooplankton and for colonization of the marine pelagial. The divergence of the Pacific clade most likely was driven by the adaptation to this novel ecological niche rather than by geographic isolation.
Keywords: Phaeobacter gallaeciensis; attached bacteria; bacterial adaptation; genome evolution; high-throughput cultivation; zooplankton.
Figures



References
-
- Alvain S., Moulin C., Dandonneau Y., Loisel H. (2008). Seasonal distribution and succession of dominant phytoplankton groups in the global ocean: a satellite view. Glob. Biogeochem. Cycles 22:GB3001 10.1029/2007GB003154 - DOI
-
- Badewien T. H., Winkler H., Arndt K. L., Simon M. (2016). Physical Oceanography during SONNE Cruise SO248 (BacGeoPac). Oldenburg: University of Oldenburg; 10.1594/PANGAEA.864673 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

